* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Primer design - ILRI Research Computing
DNA sequencing wikipedia , lookup
DNA barcoding wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Molecular evolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Homology modeling wikipedia , lookup
Deoxyribozyme wikipedia , lookup
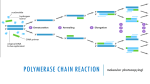
Primer design Introduction: PCR (Polymerase Chain Reaction) is a revolutionary method developed by Kary Mullis in the 1980s. PCR is based on using the ability of DNA polymerase to synthesize new strand of DNA complementary to the offered template strand. This makes it possible to delineate a specific region of template sequence that the researcher wants to amplify. At the end of a PCR reaction, the specific sequence will be accumulated in billions of copies (amplicons). Pairs of primers should have similar melting temperatures since annealing in a PCR occurs for both simultaneously. • A primer with a Tm (melting temperature) significantly higher than the reaction's annealing temperature may mishybridize and extend at an incorrect location along the DNA sequence, • Tm significantly lower than the annealing temperature may fail to anneal and extend at all. Primer sequences need to be chosen to uniquely select for a region of DNA, avoiding the possibility of mishybridization to a similar sequence nearby. A commonly used method is BLAST search whereby all the possible regions to which a primer may bind can be seen. Both the nucleotide sequence as well as the primer itself can be BLAST searched. The free NCBI tool Primer-BLAST integrates primer design and BLAST search into one application. Many online tools are freely available for primer design, some of which focus on specific applications of PCR. The popular tools Primer3Plus and PrimerQ uest can be used to find primers matching a wide variety of specifications. Considerations 1. Mononucleotide and dinucleotide repeats should be avoided, as loop formation can occur and contribute to mishybridization. 2. Primers should not easily anneal with other primers in the mixture (either other copies of same or the reverse direction primer); this phenomenon can lead to the production of 'primer dimer' products contaminating the mixture. 3. Primers should also not anneal strongly to themselves, as internal hairpins and loops could hinder the annealing with the template DNA. It is crucial to pay attention to the orientation in which the primer binds the target DNA. The reverse primer has to be the reverse complement of the given cDNA sequence. Degenerate primers Main article: Degenerate bases Degenerate primers: Degenerate primers are actually mixtures of similar, but not identical primers. • They may be convenient if the same gene is to be amplified from different organisms, as the genes themselves are probably similar but not identical. • The other use for degenerate prss Degenerate primers are widely used and extremely useful in the field of microbial ecology. They allow for the amplification of genes from thus far uncultivated microorganisms or allow the recovery of genes from organisms where genomic information is not available. Usually, degenerate primers are designed by aligning gene sequencing found in GenBank. Differences among sequences are accounted for by using IUPAC degeneracies for individual bases. PCR primers are then synthesized as a mixture of primers corresponding to all permutations. Primer design using PRIMER BLAST When you have a target sequence / region of interest (DNA), and you want to create a set of primer pairs: 1. Go to the Primer BLAST page on ncbi (http://www.ncbi.nlm.nih.gov/tools/primer-blast/ ). 2. Enter your target sequence in FASTA format or an accession number of an NCBI nucleotide sequence in the PCR Template section of the form Fasta sequence is a way of representing a sequence (protein or DNA). A sequence in FASTA format begins with a “>” and a single-line description, followed by lines of sequence data. Eg: >My_new_sequence AGGTGCAGGGGTCTCCACGTTTTAAACACACGTTCTTCCGACGGTACTGTTTCTGCCTG CTCATGACTAAGAGGGCGCCCCGTGCAGAGGTCACCCCTAGAGGAGGTAATTTATGTAC ACAAGACATAGCCTATCTGTTACATATTTTCAGCAGCCAGGGAGCCGAACGATTTGTAG ACCTTCATCTATTACTCCCAGCCCGAGTGCCTCCATCGGTTGGTAAAAAAATCTAGGAA GTGGCGTTCGCACGGCAATCCGAGTCGAAGAGACTATTCTGTGCTGAAAAAATAATATA ATCTCTGACCACATCCCATCGGCAGCTTGTGCAAGGTAGGTTCATTACGTAACTAGCTA CCGTATTTTTTACGTCAAAGTTAAATATCATCTCCGTCTTAGCGTAATCGTTGAGCTCA ACGCCTTCTACCGGCTGGAGTGAAGGGGCTACACAATTCTAACCGGACGAGGGACCCCC ATGATCTATCCGTTTGCCTGACGCATGGTATACGGCGACTGGAGACCTCGGAATTGATG TTGTGTGCGGAAAACCACATATGCACGGGCTACCCCGATCTATCAGTCCATGGTCATCA GATAGTATATCGGTACTGAACCCTAGGTGAGAATAACCTCGCGGTTTCTCTGAACTAAC GGCTTATCTGTTTGTCCGTGGTCATCCCGGGATGGTCGTGGGACATGGCACAGCCTAAC CAGCATCATAACTAGTTGAAATATTGGCTGTCTTTGTGATAAACAGTTATAAGTCATCT ATGGGGGGCGACCTACATAGTAAGTCAAAGTTTCCTACACCTTTCTACTGAGTGTATAT CCAGTTACTTGGAAGATACGCAGCGAGCCAGGTATGCTGATCACGTACTGTAGGATTAC TGGCGTATTTTCAGACGCCCCTAGAAACGTGGCTGGCCAAAGGATGATCTTGACGCCTG ACACACACGCACGATGTCCTTGAACCAGTATACCACCCTTAATTGTCTGGAGAGAT 3. If one or both primer sequences are to be used in the search, enter these in the Primer Parameters section of the form. Primer BLAST performs only a specificity check when a target template and both primers are provided. 4. In the Primer Pair Specificity Checking Parameters section, select the appropriate source Organism and the smallest Database that is likely to contain the target sequence. These settings give the most precise results. For broadest coverage, choose the nr database and do not specify an organism. Click the "Get Primers" button to submit the search and retrieve specific primer pairs Testing primer specificity for primers designed using alternative methods (such as CLC) If you have a primer pair(s) and you want to test whether they are specific to amplify the target DNA region, you can do so using the following steps: 1. Go to NCBI BLAST page (http://blast.ncbi.nlm.nih.gov/Blast.cgi ) 2. Paste the Forward primer first, then type about 20 “N”s without leaving spaces, and then paste the Reverse compliment of the Reverse primer. N is the IUPAC symbol for any base. Your search box should look something like this: ATGACACGGAGGTCTACGGANNNNNNNNNNNNNNNNNNNNNNNNNNCAAGTCCCAGGCTCTGTCTG 3. Under “Choose the search set” start typing the name of the organism for which you are working with, and click BLAST 4. You will get the BLAST hits for the sequence you are targeting appear as: TIPS 1. Design your PCR primers to be 18-30 oligo nucleotides in length. The longer end of this range allows higher specificity and gives you space to add restriction enzyme sites to the primer end for cloning. 2. Make sure the melting temperature (Tm) of the primers used are not more than 5°C different from each other. You can calculate Tm with this formula: Tm = 4(G + C) + 2(A + T)°C 3. Aim for a Tm between 65 and 70°C for each primer over the region of hybridization 4. Use an annealing temperature (Ta) of 10 to 15°C lower than the Tm. 5. The GC content of each primer should be in the range of 40-60% for optimum PCR efficiency. 6. Try to have uniform distribution of G and C nucleotides, as clusters of G’s or C’s can cause non-specific priming. 7. Avoid long runs of the same nucleotide. 8. Check that primers are not self-complementary or complementary to the other primer in the reaction mixture, as this will encourage formation of hairpins and primer dimers and will compete with the template for the use of primer and reagent. 9. If you can, make the 3′ end terminate in C or A, as the 3′ is the end which extends and neither the C or A nucleotide wobbles. This will increases the specificity. 10. You can avoid mispriming by making the 3′ end slightly AT rich.