* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Image Analysis primer - The Rutgers
Survey
Document related concepts
Transcript
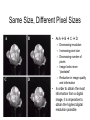
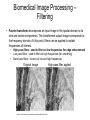
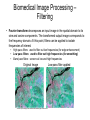
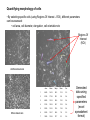
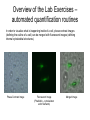
Cellular Bioengineering Boot Camp Image Analysis Overview of the Lab Exercises Microscopy and Cellular Imaging The purpose of this laboratory exercise is to develop an understanding of the measurements of cellular property via biomedical imaging and microscopy techniques. After completing this laboratory experiment, you should be able to: 1. Obtain total cell population and percentage dead cells and quantify on the parameter basis of color and cell diameter using the ImageJ software. 2. Quantify, compare and relate different morphological cell configurations to function. Electromagnetic Spectrum The wavelength required to “see” an object must be the same size or smaller than the object Cells ~10-100 um, Larger than visible light wavelengths (400-800 nm) Source: Wikipedia Biomedical Image Processing - Pixels Source: http://hosting.soonet.ca/eliris/remotesensing/bl130lec10.html Images are comprised of pixels: 2-D Picture Elements. Each pixel contains intensity information: Gray-scale Intensity (0-256 for 8-bit) 0 is black, 1 is white Same Size, Different Pixel Sizes A C B D • As A B C D: – Decreasing resolution – Increasing pixel size – Decreasing number of pixels – Image looks more “pixelated” – Reduction in image quality and information • In order to obtain the most information from a digital image, it is imperative to obtain the highest digital resolution possible Biomedical Image Processing – Segmentation & Thresholding • A gray level histogram - graphical representation of the grayscale levels that comprise an image. • Can set thresholds as desired, ex: red line in the histogram below to create binary images (all pixels either 0 or 1) – – Reduces data storage Images easier to analyze Source: http://www.olympusmicro.com/primer/java/digitalimaging/processing/automaticthresholding/ Biomedical Image Processing – Filtering • Fourier transform decomposes an input image in the spatial domain to its sine and cosine components. The transformed output image corresponds to the frequency domain. At this point, filters can be applied to isolate frequencies of interest. • • • High pass filters: used to filter out low frequencies (for edge enhancement) Low pass filters: used to filter out high frequencies (for smoothing) Band pass filters: screen out low and high frequencies Original Image Source: http://hosting.soonet.ca/eliris/remotesensing/bl130lec10.html High-pass filter applied Biomedical Image Processing – Filtering • Fourier transform decomposes an input image in the spatial domain to its sine and cosine components. The transformed output image corresponds to the frequency domain. At this point, filters can be applied to isolate frequencies of interest • • • High pass filters: used to filter out low frequencies (for edge enhancement) Low pass filters: used to filter out high frequencies (for smoothing) Band pass filters: screen out low and high frequencies Original Image Source: http://hosting.soonet.ca/eliris/remotesensing/bl130lec10.html Low-pass filter applied Overview of the Lab Exercises – automated quantification routines Cells can be selected & quantified according to different parameters • cell physical characteristics (diameter, area, elongation etc) • cell fluorescence intensity Diameter measurement Cell recognition using fluorescence intensity Criteria (thresholds) ID Particle Mean Green ID Particle Mean Green 1 211 7 225 2 209 8 222 3 170 9 205 4 219 10 210 5 213 11 214 6 196 12 222 ImageJ generated fluorescence intensity values (excel spreadsheet format) Software cell recognition using diameter criteria Quantifying morphology of cells • By selecting specific cells (using Regions Of Interest – ROI), different parameters can be assessed: • cell area, cell diameter, elongation, cell orientation etc Regions Of Interest (ROI) Undifferentiated cells Differentiated cells Area Mean Major Minor Circ. 1 604.0 154.3 62.3 12.3 0.3 2 218.0 173.8 29.1 9.5 0.4 3 496.0 153.3 52.6 12.0 0.4 4 315.0 155.9 36.7 10.9 0.4 5 236.0 161.6 21.8 13.8 0.4 6 263.0 163.7 42.8 7.8 0.3 7 198.0 153.8 27.7 9.1 0.6 Generated data using specified parameters (excel spreadsheet format) Overview of the Lab Exercises – automated quantification routines In order to visualize what is happening inside of a cell, phase contrast images (defining the outline of a cell) can be merged with fluorescent images (defining internal cytoskeletal structures). Phase Contrast Image Fluorescent Image (Phalloidin – cytoskeleton actin filaments) Merged Image Example of Biomedical Image Processing • Biomedical polymer sponges were saturated with a fluorophore (fluorescein isothiocyanate - FITC) & imaged on a microscope using fluorescence microscopy. • Images of each porous field were digitized and stored on the computer. • Digitized images were analyzed using biomedical image processing to identify the number, shape, and location of pores. (Similar approaches can be used to identify fluorescently labeled cells and cell number, cell viability, cell morphology) 3. Open 1. Shading Correction 2. Segmentation 4. Scrap & Fill 5. Measurements Tjia and Moghe, J. Biomed. Mater. Res (Appl Biomat) 43: 291, 1998