* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Asgeirsson, B., Renzetti, G., Invernizzi, G ., Papaleo, E. (2013)
Intrinsically disordered proteins wikipedia , lookup
Protein domain wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein structure prediction wikipedia , lookup
Homology modeling wikipedia , lookup
Alpha helix wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Structural alignment wikipedia , lookup
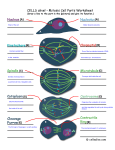
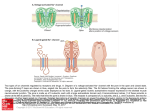
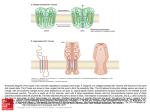
Asymmetric flexibility of a homodimeric enzyme as shown by molecular dynamics computations. A case study of the cold-active Vibrio alkaline phosphatase. Ásgeirsson, B.1, Renzetti, G.2, Invernizzi, G.2, Papaleo, E.2 2 1Science Institute, University of Iceland, Dunhaga 3, 107 Reykjavik, Iceland. Department of Biotechnology and Biosciences, University of Milano-Bicocca, Piazza della Scienza 2, 20126, Milan, Italy. Introduc4on Multiple all-atom explicit solvent molecular dynamics simulations were employed in conjunction with different metrics to analyze the dynamics patterns and the paths of intra- and intermolecular communication in a cold-active alkaline phosphatase (VAP)1. Asymmetric dynamics have been suggested to play a part in the catalytic cycle in homodimeric alkaline phosphatases. A conformational change might be the rate-limiting step since the chemical transformations are much faster than kcat2. Asymmetric protein dynamics would influence protein function and stability by modulating conformational changes consistent with half-ofthe-sites mechanism3,4. Here, we wanted to see if the symmetric crystal structure of VAP would become asymmetrical in solution, a good predictor of half-of the-sites mechanism. Results Fig.1 - Dynamic patterns of the two subunits had a different distribution of intramolecular interactions and correlated motions (rmsf). Fig. 2 - VAP displayed a low number of intersubunit interactions. Coupled motions between the two halves were also few. Fig. 3 - Numerous salt-bridge clusters were observed with asymmetric distribution in the two subunits . Fig. 4 - Hub residues (those that link 3 or more other residues) were nonsymmetrically distributed. Several were located in area specific for VAP, i.e. the large loop (insert II). Asymmetric flexibility paLerns Cα correla4ons Fig. 2. Significant posi4ve correla4ons between Cα atoms. The posi=ve correla=ons are shown as green s=cks connec=ng the Cα atoms of the average structures from the simula=ons. The two subunits A and B are shown as white and pale‐cyan and pink cartoon, respec=vely, with regions of differen=al flexibility highlighted in red and blue according to Fig. 2. The cataly=c residue S65 and the metal ions Mg2+ and Zn2+ are shown as s=cks, a black sphere and grey spheres, respec=vely in both the subunits. The panel A and B present two different orienta=on of the 3D structure for sake of clarity. Salt‐bridge clusters Fig. 3. Salt‐bridge clusters in VAP. (A) The salt‐bridges are mapped on the 3D average structure from simula=ons as s=cks connec=ng Cα atoms of VAP. The shade of colors indicate the persistence degree of the salt‐bridge interac=ons from blue (high persistence) to light‐green (low persistence). The black dots indicate the loca=on of the metal ions. The different clusters of spa=al proximity of the salt‐bridges and their networks are indicated by different colors and for sake of clarity only the most populated clusters are indicated by labels. (B‐C) The intramolecular salt‐bridges and heir networks have been mapped on the individual subunit aXer structural alignment of the subunits in order to compare their distribu=on on the 3D structure of subunit A (panel B) and B (panel C). The different shade of colors of the residues involved in the salt‐ bridges, which are represented as spheres corresponding to the Cα posi=on, are only for sake of clarity and are not related to clustering of the interac=ons. Hub residues Fig. 4. Hub residues derived from Protein Structure Network analyzes of VAP MD trajectories are shown as magenta and blue spheres for subunit A and B, respec=vely. The four inserts (I to IV) not conserved in the warm‐adapted counterparts are in green. R129 and S65 are shown as s=cks. Conclusions • Dynamic patterns of the two VAP subunits were asymmetric. • VAP subunits had a different distribution of intramolecular interactions and correlated motions. • VAP displayed a low number of intersubunit interactions. Coupled motions between the two halves were also few. Figure 1. (A) The average rmsf per‐residue profiles calculated over different =me windows (whole trajectory, 10 ns, or 5 ns) of subunit A (purple shade of colors) and B (blue shade of colors) are shown. (B) The =me‐dependent rmsf profiles calculated on =me‐windows of 3 ns of subunit A (purple shade of colors) and B (blue shade of colors) show the progressive changes in residue mobili=es. (C) Regions characterized by differen=al flexibility in subunit A and B are mapped on the 3D structure. In par=cular, regions characterized by the highest flexibility in monomer A or in monomer B are shown in red and blue, respec=vely. The two subunits A and B are shown as white and pale‐cyan and pink cartoon, respec=vely. The four inser=ons characteris=cs of VAP are highlighted by different colors in both the subunits. The cataly=c residue S65 and the metal ions Mg2+ and Zn2+ are shown as s=cks, a black sphere and grey spheres, respec=vely in both the subunits. (D‐E). The rmsf profiles have been mapped on the 3D structure of monomer A(D) and B (E) aXer a structural alignment of the two subunits. In par=cular, cartoon shade of color and thickness are propor=onal to the Cα rmsf values. [email protected] [email protected] Our results provide a structural rationale to support the half-of-thesites mechanism for VAP. This will help us to understand the reaction mechanism of this metallo-phosphatase better and open the door for studying how cold-adaptation has generated a cold–active variant by mutagenesis. References (1) Papaleo E, Renze] G, Invernizzi G, Ásgeirsson B. Dynamics fingerprint and inherent asymmetric flexibility of a cold‐ adapted homodimeric enzyme. A case study of the Vibrio alkaline phosphatase. Biochim Biophys Acta (BBA) ‐ General Subjects. 2013;1830(4):2970‐2980. (2) Halford, S. E., Bennett, N. G., Trentham, D. R., and Gutfeund, H. (1969) A substate-induced conformation change in the reaction of alkaline phosphatase from Escherichia coli, Biochem J 114, 243-251. (3) Chappelet-Tordo, D., Fosset, M., Iwatsubo, M., Gache, C., and Lazdunski, M. (1974) Intestinal alkaline phosphatase. Catalytic properties and half of the sites reactivity, Biochemistry 13, 1788-1794. (4) Orhanovic, S., and Pavela-Vrancic, M. (2003) Dimer asymmetry and the catalytic cycle of alkaline phosphatase from Escherichia coli, Eur J Biochem 270, 4356-4364. Acknowledgement: The Icelandic Centre for Research and University of Iceland provided financial support