* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 15, Feb 26
Butyric acid wikipedia , lookup
Citric acid cycle wikipedia , lookup
List of types of proteins wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Self-assembling peptide wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Bottromycin wikipedia , lookup
Genetic code wikipedia , lookup
Peptide synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
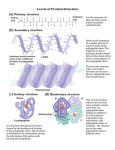
BIO 311C Spring 2010 Lecture 15 – Friday 26 Feb. 1 Review Illustration of a Polypeptide amino acids peptide bonds Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration 3 Folding and other modifications Functional Protein * Structural Formula of an Amino Acid A single amino acid that is not chemically attached to anything else is called a "free amino acid". central carbon atom Characteristics of Free Amino Acids A central carbon atom is attached to four chemical groups. The central carbon is an asymmetric carbon atom unless R is identical to one of the other three chemical groups. Both the amino and the carboxylic acid functional group of free amino acids are ionized at near neutral pH. 4 * Ways of Showing the Structural Formula of an Amino Acid Uncharged form; does not occur in aqueous solutions at neutral pH. Charged form that occurs in aqueous solutions at near neutral pH. Detailed structure 5 Abbreviated Structure * Amino Acids Used to Construct Proteins central carbon atom H + H3N C COO Twenty different kinds of amino acids are used for constructing proteins, each with a different "R" group. R Since the central carbon atom of an amino acid is asymmetrical, there are 2 enantiomers of each kind of amino acid. Cells use only the "L" enantiomer of each kind of amino acid to construct proteins. The R-group of a few kinds of amino acids carry an amino or a carboxylic acid functional group. These amino acids carry three electrical charges at pH values near 7. 6 * Aspartic Acid Glycine (shown without its (shown without its functional groups ionized) functional groups ionized) Threonine (shown without its functional groups ionized) How many electric charges would this free amino acid have in the cytoplasmic matrix of a living cell? How many electric charges would this free amino acid have in the cytoplasmic matrix of a living cell? How many electric charges would this free amino acid have in the cytoplasmic matrix of a living cell? How many asymmetric carbon atoms does this free amino acid contain? How many asymmetric carbon atoms does this free amino acid contain? How many asymmetric carbon atoms does this free amino acid contain? 7 * Formation of a Peptide Bond from Two Amino Acids These atoms are lost as a molecule of water during the formation of a peptide bond. These atoms are covalently bonded together during formation of a peptide bond In most cases R1 and R2 will not be the same chemical structures. 8 * Dipeptide Formation From Two Amino Acids + H2O aa1 + aa2 dehydration reaction dipeptide + H2O Atoms of peptide bond 9 * A Peptide Bond Between Two Amino Acids A peptide bond between two amino acids consists of the four atoms shown at right. All 6 atoms shaded in grey (above) lie in the same rigid plane, with the double-bonded oxygen projecting in the opposite direction from the hydrogen atom that is bonded to the nitrogen atom. O C The carboxylic acid and amino functional groups that formed the peptide bond (see previous slide) can no longer ionize after they have become incorporated into a peptide bond. 10 N H * Oligopeptides and Polypeptides dipeptide: Two amino acids covalently bonded together by a peptide bond: oligopeptide: 3 - 20 amino acids covalently bonded together by peptide bonds tripeptide: Three amino acids covalently bonded together by two peptide bonds tetrapeptide: Four amino acids covalently bonded together by three peptide bonds polypeptide (also called a polypeptide chain): More than twenty amino acids covalently bonded together by peptide bonds 11 * Oligopeptides and polypeptides have two distinct ends: - an N-terminal end (also called an amino-terminal end, - a C-terminal end (also called a carboxy-terminal end). Example N-terminal end + H3N H O C C R1 H H N H C R2 C N C-terminal end COO - O A tripeptide Can you identify the peptide bonds in this molecule? 12 * Hydrogen Bonding Between Two Peptide Groups Direction of polypeptide chain N-terminal end hydrogen bond C-terminal end Direction of polypeptide chain The structure imposed on portions of a polypeptide chain due to hydrogen bonding between different peptide bonds is called the secondary structure of the polypeptide chain. 13 * Multiple hydrogen bonds can form between adjacent peptide groups along the folded polypeptide chain Direction of polypeptide chain N-terminal end additional hydrogen bond hydrogen bond C-terminal end Direction of polypeptide chain 14 * A common secondary structure of polypeptide chains resembles the shape of a stretched spring and is called the alpha-helix conformation. Abbreviated illustration showing back-bone structure and ribbon diagram of a portion of a polypeptide chain in an α-helix conformation. Illustration showing all atoms of a portion of a polypeptide chain in an α-helix conformation. Yellow lines represent hydrogen bonds. This conformation is stabilized by multiple hydrogen bonds between atoms of different peptide bonds. 15 * Another common secondary structure of polypeptide chains resembles the shape of a sheet of paper folded back and forth; it is called the betasheet (or pleated sheet or beta pleated sheet) conformation. The polypeptide chain may fold back again to allow three (or more) regions of the polypeptide chain to align in a thicker pleated sheet conformation. Note the oxygen and hydrogen atoms of peptide bonds that are projecting out such that they are available to form additional hydrogen bonds, which would expand the pleated sheet. The R groups actually project at approximately right angles to the plane of the pleated sheet. Illustration showing back-bone structure and diagram of two portions of a polypeptide chain that are aligned together in a pleated sheet conformation. As with the alpha-helix conformation, the pleated-sheet conformation is stabilized by hydrogen bonding between atoms of different peptide bonds. 16 * Categories of R Groups that Occur in Amino Acids I. No R-group (glycine contains only H) II. With R-group A. Nonpolar R-Group B. Polar R-Group 1. non-ionized 2. ionized a. cationic b. anionic 18 generalized amino acid * Leucine Serine an amino acid with a nonpolar R-group an amino acid with a polar, non-charged R-group R-groups are shown with a light pink background. 19 * Lysine Aspartic acid an amino acid with a cationic R-group an amino acid with an anionic R-group R-groups are shown with a light pink background. 20 * The Amino Acid Sequence of a Polypeptide Chain From textbook Fig. 5.21, p. 82 Each living cell produces several thousands of different kinds of polypeptide chains. Some Features of This Specific Polypeptide Chain It consists of 127 amino acids. Its N-terminal amino acid is Gly (G). Its C-terminal amino acid is Glu (E). Amino acid number 25 is Ala (A). Each kind of polypeptide chain has an exact number of amino acids. The sequence of amino acids is exactly the same for each copy of the same kind of polypeptide chain. The amino acids of a polypeptide chain are numbered sequentially, starting from the N-terminal (aminoterminal) end. 21 * Representation of a the Primary Structure of a Portion of a Specific Polypeptide Chain amino acids peptide bonds - + H3N COO Ala A Asp D Gly G Phe F Leu L Lys K Ser S Cys C Dotted line indicates that the polypeptide chain continues. Dotted line indicates that the polypeptide chain continues. Previously amino acids were given three-letter abbreviated names; Now they are usually designated by one-letter abbreviated names. The specific sequence of amino acids along a polypeptide chain, starting at the N-terminal end, is called the primary structure of the polypeptide chain. 22 In general the sequence is not a regular, repeating pattern. It is unique to each different kind of polypeptide chain. * Secondary Structures Contribute to the Overall Size and Shape of a Polypeptide Chain From Textbook Fig. 5.19, p. 81 An alpha-helix secondary structure A beta-sheet secondary structure Each alpha helix and each beta (pleated) sheet contributes to only a portion of the overall threedimensional shape of the polypeptide chain. 23 * Examples of Specific R-groups Along a Region of a Polypeptide Chain one-letter amino acid identifier backbone of Polypeptide chain R-groups of amino acids The differing R-groups on different amino acids carry functional groups that confer specific properties to each amino acid positioned along the polypeptide chain. Some R groups along the polypeptide chain are nonpolar, some are polar and uncharged, some are positively charged, and some are negatively charged. 24 * Kinds of Bonds that May Form Between Different R-Groups When a Polypeptide Chain Bends and Folds in Various Ways From textbook Fig. 5.21, p 83 Covalent bond (Disulfide bond) Polar bonds Hydrogen bonds Electrovalent (ionic) bonds Hydrophobic bonding 25 * Representations of the Conformation of a Functional Polypeptide Chain From Fig. 5.19, p. 81, of textbook α helix β sheet disulfide Ribbon model Space-filling model lysozyme The three-dimensional shape (conformation) of a polypeptide chain is called its tertiary structure. 26 The secondary structure and tertiary structure of a polypeptide chain are determined by its primary structure. Thus, each different polypeptide chain with its unique sequence of amino acids also has its own unique conformation. * A ribbon structure of ribonuclease is shown, illustrating its native conformation. Ribonuclease serves as a functional enzyme when in this conformation. Ribonuclease A polypeptide chain that is folded into its normal, functional conformation is said to be in its native conformation. A polypeptide that is folded improperly so that it cannot function is said to be to be denatured or in a denatured conformation. 27 * Relationship Between a Protein and a Polypeptide Chain A polypeptide chain is a sequence of 20 or more amino acids held together by peptide bonds. When one or more polypeptide chains are folded in a conformation that forms a functional unit, then the unit is called a protein (or an active protein). Thus, all proteins contain at least one polypeptide chain. * 28 Monomeric and Oligomeric Proteins α-tubulin lysozyme is a monomeric protein β-tubulin tubulin is a dimeric protein A protein that consists of only one polypeptide chain is called a monomeric protein. Microtubules are polymeric proteins A protein that consists of several (2 or a few) polypeptide chains is called an oligomeric protein. The three-dimensional shape of an oligomeric protein is called its quaternary structure. 29 The individual polypeptide chains of an oligomeric protein are called subunits of the protein. *