* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Conservation of surface epitopes in Pseudomonas aeruginosa outer
Gene nomenclature wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Expanded genetic code wikipedia , lookup
DNA vaccination wikipedia , lookup
Pathogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Protein moonlighting wikipedia , lookup
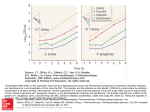
261 FEMS Microbiology Letters 113 (1993) 261-266 © 1993 Federation of European Microbiological Societies 0378-1097/93/$06.00 Published by Elsevier FEMSLE 05664 Conservation of surface epitopes in Pseudomonas aeruginosa outer membrane porin protein OprF Nancy L. Martin 4, Eileen G. Rawling 4, Rebecca S.Y. Wong and R o b e r t E.W. H a n c o c k .,a a Mac Rosok b a Department of Microbiology, University of British Columbia, Vancouver, B.C., Canada V6T 1Z3 and b Bristol Myers Squibb Pharmaceutical Research Division, 3005 First Avenue, Seattle, Washington, USA (Received 28 June 1993; revision received 11 August 1993; accepted 17 August 1993) Abstract: The outer membrane proteins of several prominent bacterial pathogens demonstrate substantial variation in their surface antigenic epitopes. To determine if this was also true for Pseudomonas aeruginosa outer membrane protein OprF, gene sequencing of a serotype 5 isolate was performed to permit comparison with the published serotype 12 oprF gene sequence. Only 16 nucleotide substitutions in the 1053 nucleotide coding region were observed; none of these changed the amino acid sequence. A panel of 10 monoclonal antibodies (mAbs) reacted with each of 46 P. aeruginosa strains representing all 17 serotype strains, 12 clinical isolates, 15 environmental isolates and 2 laboratory isolates. Between two and eight of these mAbs also reacted with proteins from representatives of the rRNA homology group I of the Pseudomonadaceae. Nine of the ten mAbs recognized surface antigenic epitopes as determined by indirect immunofluorescence techniques and their ability to opsonize P. aeruginosa for phagocytosis. These epitopes were partially masked by lipopolysaccharide side chains as revealed using a side chain-deficient mutant. It is concluded that OprF is a highly conserved protein with several conserved surface antigenic epitopes. Key words: Porin; pseudomonas aeruginosa; Outer membrane; OprF; Epitope Introduction One of the strategies utilized by successful pathogens to evade the immune response is variation of surface antigen structure. For outer membrane proteins this can involve either mutations [1], or more complex genetic events such as rearrangements promoted by frameshifts [2]. The ma- * Corresponding author. Tel.(604) 822-2682, Fax (604) 8224040. jor outer membrane proteins of Gram negative bacteria have been shown, by either monoclonal antibody (mAb) reactivity or DNA sequence analysis to vary within individual strains of many species. Thus the outer membrane porin proteins from Escherichia coli [3], Haemophilus influenzae [4], Chlamydia trachomatis [5] and Neisseria sp. [6] have all been shown to have variations in surface antigenic structure, whereas internal epitopes are generally more highly conserved. OprF is the major porin protein in P. aeruginosa that forms channels through the outer mem- 262 brane that are large enough to accommodate diand tri-saccharides [9]. In addition, a second function in both outer membrane and cell structure and stability was discovered, in that OprFdeficient mutants have rounded morphology and grow well only in high osmolarity media [10,11]. Studies utilizing mAbs specific for OprF have demonstrated that this protein is potentially useful as a target for diagnostic and immunotherapeutic intervention [12,13]. Indeed, Gilleland and colleagues [14] have presented data to demonstrate that immunization with O p r F protects against subsequent Pseudomonas aeruginosa infections. Previous studies with two mAbs directed against Pseudomonas aeruginosa protein OprF demonstrated the conservation of two surface epitopes that were present in all P. aeruginosa strains tested [7]. Furthermore, the oprF gene cross-hybridized in all strains and demonstrated rather conserved restriction patterns [8]. However, the restricted number of mAbs tested, and the observation of variation in surface antigenic structure in other bacterial pathogens, led us to examine here whether OprF was an unusual example of an outer membrane protein with general conservation of surface epitopes. Materials and Methods Strain and media P. aeruginosa PAO strains utilized included wild type strain H103 (IATS serotype 5; Hancock and Carey, 1979), its OprF::£1 mutant H636 [10] and H692, a mutant unable to produce A and B band LPS side chains (a gift from Dr. J. Lam, University of Guelph, Canada). Strain M2 [13] was an IATS serotype 5 strain (i.e., the same serotype as strain H103). Other Pseudomonadaceae strains used were described previously [7,16,17]. Growth media included Mueller Hinton Broth (all strains) and proteose peptone No. 2 (PP2) without (H103, M2) or with (H636) the addition of 200 mM NaC1 and 500 /xg/ml streptomycin. Strain H692 was grown on 1% Bactotryptone, 0.5% yeast extract, 0.5% NaC1. Immunological techniques Enzyme-linked immunoabsorbent assays were performed with wells coated with either whole cells, isolated outer membranes or purified OprF, following procedures previously described by Mutharia and Hancock [17]. Cell surface indirect immunofluorescence labelling was carried out on mid-logarithmic or stationary phase cells collected by centrifugation from 1.5 ml of culture. The cells were washed twice with PBS by centrifugation and resuspended in 200 /.tl of PBS. This suspension was smeared on a glass slide (that in some experiments was precoated with 1 m g / m l poly-L-lysine) and allowed to air dry, then fixed in 100% ethanol. The slides were then processed following the method of Hofstra et al. [18], with minor modifications as described by Mutharia and Hancock [17]. The slides were examined with a Zeiss microscope fitted with a condenser for fluorescence microscopy and containing a halogen lamp and suitable filters for emission of fluorescein isothiocyanate at 525 nm. Opsonized phagocytosis of P. aeruginosa was performed using a modification of the procedures of Battershill et al. [19]. Mouse peritoneal macrophages from 2 - 4 months old B6D2(fl) mice were incubated in 8-well chamber slides (NUNC, Naperville, IL) overnight and rinsed twice prior to addition of bacteria, thereby eliminating the cytospin step. Antibody concentrations used were standardized by ELISA with purified OprF. Experiments were carried out on 3 different days, the results from each day were tabulated and subjected separately to the Mann-Whitney test to determine a significant difference from the control (cells opsonized with mAb MA1-3). Colony immunoblotting [7] and Western immunoblotting [17] were carried out as described previously. DNA sequencing The oprF gene of P. aeruginosa strain H103 [11] was sequenced as described previously using oligonucleotide primers [8], except that the protocols of Applied Biosystems Inc. (ABI) were followed for sequencing with an ABI Model 373 automated fluorescence DNA sequenator. The 263 the oprF gene from a serotype 12 isolate that had been sequenced (Genbank accession number P13794) by Duchene et al. [20], whereas the majority of strains, including our laboratory wild type strain H103 and strain M2, lacked this Kpnl site. To determine the extent of variation of oprF sequences in P. aeruginosa, the cloned gene [11] from strain H103 was sequenced in this study (Genbank accession number M94078). Only 16 nucleotide pair changes were observed relative to the serotype 12 sequence. Nine of these were C --) T (at nucleotide positions 69, 175, 225, 486, 624, 650, 789, 954 and 1,029 of the open reading frame), four were T---> C (at positions 228, 267, 516, and 837), two were T --) G (at positions 150 and 204), and one was G--) A (at position 930). All of these changes were silent and thus the two amino acid sequences were identical. MA7-4,5,7 MA7-2 MA7-3 MA7-1 MA4-4,MA7-8 M I M I M l I__ 50 7-----150 100 F i g . 1. L i n e a r m a p o f O p r F approximate topes based positions on data from MAS-8 M I [ I 200 I 250 I 300 with positions of the methionine (M) and cysteine (C) residues the CCCC ~lf MA7-6 indicated. of the N. Martin, 1 9 9 2 a n d [15l. T h e n u m b e r s Above monoclonal the map are antibody Ph.D. epi- Thesis U.B.C., refer to amino acid positions. D N A sequence is deposited in Genbank under the accession number M94078. Results Antigenic conservation of OprF in Pseudomonas aeruginosa Sequence conservation of OprF in Pseudomonas aeruginosa A panel [15] of ten mAbs recognizing diverse epitopes of OprF (Fig. 1) were reacted with a wide variety of P. aeruginosa strains using the colony immunoblot procedure [7]. All isolates of P. aeruginosa, except for the negative control strain H636 oprF::O, reacted on colony immunoblots with all 10 mAbs (Table 1), as confirmed in selected cases by Western immunoblot- Previous experiments [8] probing, by Southern hybridization, the oprF gene of 59 P. aeruginosa clinical and serotyping strains indicated only one restriction endonuclease polymorphism observed in 2 strains which demonstrated an additional K.pnl site in the carboxy terminal half of the oprF gene. This unusual pattern was also observed for Table 1 Monoclonal antibody reactivity of various Pscudomonads Strains a a The designation for homology group I Reactivity b P. aeruginosa ( 4 6 i s o l a t e s ) P. syringae A T C C 1 9 3 1 0 x P. fluoresccens A T C C 135253P.fluorescens O E 2 8 . 3 P. putida A T C C 126333P. stutzerii A T C C 1 7 5 7 8 T P. aureofaciens A T C C 139853P. chloraphis A T C C 9 4 4 6 T A. vinelandii O P b + + Reaction of rRNA superscript on Western P. aeruginosa OprF; MA4-4 MAS-8 MA7-1 MA7-2 MA7-3 MA7-4 MA7-5 MA7-6 MA7-7 + + + + + + + + + + + + + + + + + + + + + + - - + + - - - + + - + + + - - - + + - _ _ + + _ + _ + + + + _ + + - - + + + + + + + + + + + - - + + + - - + + + + - - - + + + - + + - + + + - - - + + + - + + - + + + - - - - + + - - - + - . _ c . . T i n d i c a t e s t h a t t h i s is t h e t y p e s t r a i n o f t h e s p e c i e s . immunoblot - . MA7-8 equivalent to OprF of no reaction. All samples P. aeruginosa were heated strain H692; + reaction of lesser intensity than that a t 1 0 0 ° C f o r 15 m i n in s o l u b i l i z a t i o n 2-mercaptoethanol, prior to SDS-PAGE. R e s u l t s f o r t h e 46 P. aeruginosa s t r a i n s w e r e o b t a i n e d v a l i d a t e d in s e l e c t e d i n s t a n c e s b y W e s t e r n i m m u n o b l o t . c Reactivity was observed with a band of lower apparent molecular mass. reduction mix without by colony immunoblot [7l a n d 264 ting and ELISA studies. The P. aeruginosa strains tested included the 17 IATS serotype isolates [17], 15 clinical strains from different countries, 15 environmental isolates, and 3 laboratory isolates. These data thus indicated strong conservation of OprF epitopes in P. aeruginosa. Table 2 Indirect immunofluorescence labelling of intact P. aeruginosa by O p r F - s p e c i f i c m o n o c l o n a l a n t i b o d i e s a n d t h e c o n t r o l a n t i body MAI-3 Monoclonal antibody lmmunofluorescence M2 H103 " H692 MA4-4 + + + + + + Reactivity with other Pseudomonadaceae Southern blotting studies with the oprF gene from P. aeruginosa previously indicated that the oprF gene was conserved in all Pseudomonadaceae of rRNA homology group I [8]. Two MA5-8 + + - + + MA7-1 + + - + MA7-5 additional genes have now been sequenced, the P. syringae pv. syringae ATCC 19310 OprF porin [8] and the P. fluorescens OE283 plant root adhesion protein [21]. Interestingly, although both have extensive amino acid homology with P. aeruginosa OprF, the latter lacks the entire cysteinecontaining region of OprF, whereas the former contains this region [21]. Consistent with this, P. syringae OprF reacted with mAbs MA4-4 and MA7-8, which recognize the cysteine disulphide region [15], whereas P. fluorescens OE283 did not (Table 1). Of the Pseudomonadaceae strains examined by Western immunoblot analysis (Table 1), P. putida showed the broadest cross-reactivity of any of the strains examined here, reacting with 8 of the 10 mAbs. However, all strains examined reacted with 2 or more mAbs. The weakest cross-reactivity was observed with ATCC 13525, the type strain of P. fluorescens, and with Azotobacter vinelandii. Three mAbs MA7-2, MA7-6 and MA7-7 represented the most conserved epitopes reacting with 7, 7 and 8 strains, respectively. Conversely, the epitopes for MA5-8, MA7-5 and MA7-1 were poorly conserved demonstrating weak reactivity with 0 to 2 other Pseudomonads. Surface localization studies. It has been previously demonstrated that surface localized regions of outer membrane proteins demonstrate the greatest variability [3,4,22]. Thus, it was possible that the antigenic conservation described above reflected conservation of epitopes that were not exposed on the surface. Therefore, indirect immunfluorescence studies were performed to determine whether or not the OprF epitopes being studied were surface localized (Table 2). Utilizing MA7-6 MA7-7 MA7-8 MA1-3 b H636 - MA7-2 - - + MA7-3 + + + + - MA7-4 + + + + + - + + + + - + - + + + + + + - + + - + + NT c NT a _ - No fluorescence or only weak fluorescence observed, + cells u n i f o r m l y fluorescent, + + cells highly fluorescent. b Negative control antibody specific for a non surface-localized epitope on outer membrane protein OprI. " NT, not tested. strain M2, we observed labelling of the surfaces of cells with all antibodies studied except MA7-2 and the negative control [19] antibody MA1-3. Labelling of strain H103 was consistently weaker and several antibodies failed to label strain H103. This appeared to be due in part to masking of OprF by LPS side chains since strain H692, an LPS-altered, rough derivative of strain H103 was better labelled with all mAbs. Strain H636, an oprF::~2 mutant of strain H103, was not labelled by any of the mAbs. As an alternative method of studying surface binding, each of the mAbs was utilized as an opsonin for phagocytosis of strain M2 by nonelicited mouse peritoneal macrophages. We confirmed previous data [19] which suggested that MA5-8 and MA4-4 were opsonic and that MA5-8 was superior to MA4-4 in opsonizing for phagocytosis (Fig. 2). At an antibody dilution of 10 2 MA7-3, MA7-5, MA7-6, MA7-7 and MA7-8 also consistently opsonized P. aeruginosa for phagocytosis. MAT-2 was no better than the control antibody MA1-3 whereas MA7-1 and MA7-4 resulted in a significant increase in phagocytosis in 1 out of 3 experiments. When less diluted (10 -1) antibody was also utilized for phagocytosis, MA7-1 consistently gave significant phagocytosis, while 265 ¢ a 3 .... * * : ~- : ili! 2 * (0 . * : ~ * ro 0 7-1 7-2 7-3 7-4 7-5 7-6 7-7 7-8 4-4 5-8 Antibody Fig. 2. E n h a n c e m e n t of phagocytosis of P. aeruginosa strain M2 by OprF-specific monoclonal antibodies. Results represent the average n u m b e r of bacteria associated per mouse peritoneal macrophage. Each different shaded bar represents data from an independent experiment with the control number of bacteria per macrophage (obtained using the negative control antibody MA1-3) subtracted. The star indicates samples that were significantly different from the control (by M a n n - W h i t n e y test P < 0.05). other antibodies yielded the same types of results as shown in Fig. 2. Overall the data suggested that the epitope for MA7-2 is not exposed (except perhaps in the LPS-altered strain H692). However, all other epitopes were apparently surface exposed. Discussion In contrast to several outer membrane proteins found in other bacterial species [2-6], OprF was demonstrated here to contain a number of discrete, highly conserved surface antigenic epitopes. The mAbs utilized in this study recognized epitopes that were largely clustered in the carboxy terminal half of the protein. Nevertheless, a variety of data including their variable reactivity with OprF equivalents from other Pseudomonads (Table 1) and their differential reactivity with peptides [15; N. Martin, Ph.D. Thesis, U.B.C., 1992] indicate that they recognized separate epitopes. The reason for conservation of OprF epitopes was not definitively determined in this study. In other bacteria, it has been suggested that masking of outer membrane protein surface epitopes by LPS limits their reactivity with antibod- ies specific for these epitopes [23]. This also appeared evident in this study comparing strain H103 with its LPS-altered, rough derivative H692 (Table 2). However, this cannot be the only important factor since immunofluorescence labelling of strain H103 was observed with some mAbs and one of those mAbs MA4-4 [13] protects mice against Pseudomonas infections. Furthermore, cystic fibrosis patient isolates, that demonstrate rough LPS, were fully reactive with all 10 mAbs, indicating that these isolates had not undergone extensive oprF gene mutations. Pseudomonas species from rRNA homology group I demonstrated immunologic cross-reactivity with subsets of the 10 OprF-specific mAbs, indicating that OprF has been conserved across species barriers. Three of the Pseudomonas species tested have now had their oprF genes sequenced. Alignment of the sequences of the OprF proteins from P. aeruginosa [20], P. syringae [8] and P. fluorescens [21] revealed that the carboxy-terminal 121 amino acids are highly conserved. Compared to P. aeruginosa, P. syringae OprF has 101 identical amino acids, 12 conservative substitutions and 8 differences while P. fluorescens OprF [21] has 1 single amino acid deletion, 13 different residues, 19 conservative substitutions and 87 identical amino acids (providing the cysteine disulphide region is excluded from this analysis). Only two (MA7-2, MA7-6) of the 7 mAbs specific for this region of OprF, crossreacted strongly with all 3 proteins, mAb MA7-2 did not apparently recognize an epitope that is well exposed on the surface of P. aeruginosa (Fig. 1). Thus, it seems reasonable to conclude that in these different species, the surface-localized regions, recognized by 5 of the other 6 mAbs with epitopes in the carboxy terminal 121 amino acids, have undergone the greatest antigenic variation. Consistent with this idea, the surface epitopes of porin I from Neisseria [24] and FepA from Enterobacteriaceae [22] represent the most variable portions of these proteins. Acknowledgements Financial support for this project came from the Medical Research Council of Canada. E.R. 266 and R.W. were supported by studentships from the Canadian Cystic Fibrosis Foundation and Natural Sciences and Engineering Research Council of Canada, respectively. References 1 Jeanteur, D., Lakey, J.H. and Pattus, F. (1991) The bacterial porin superfamily: Sequence alignment and structure prediction. Mol. Microbiol. 5, 2153-2164. 2 Murphy, G.L., Connell, T.D., Barritt, D.S., Doomey, M. and Cannon, J.G. (1989) Phase variation of gonococcal protein II: regulation of gene expression by slipped strand mispairing of a repetitive DNA sequence. Cell 56, 539-547. 3 Pages, J.M., Pages, C., Bernadac, A. and Prince, P. (1988) Immunological evidence for differences in the exposed regions of OmpF porins from Escherichia coli B and K-12. Molec. Immunol. 25, 555-563. 4 Van Alphen, L., Eijk, P., Van Den Brock, L.G. and Dankert, J. (1991) Immunochemical characterization of vaiable epitopes of outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 59, 247252. 5 Zhong, G. and Brunham, R.C. (1990) Immunoaccessible peptide sequences of the major outer membrane protein from Chlamydia trachomatis serovar C. Infect. Immun. 58, 3438-3441. 6 Virji, M., Fletcher, J.N., Zak, K. and Heckels, J.E. (1987) The potential protective effect of monoclonal antibodies to gonococcal outer membrane protein IA. J. Geu. Microbiol. 133, 2639-2646. 7 Mutharia, L.M. and Hancock, R.E.W. (1985) Characterization of two surface-localized antigenic sites of porin protein F of Pseudomona aeruginosa. Can. J. Microbiol. 31,381-386. 8 Ullstrom, C,A,, Siehne[, R., Woodruff, W., Steinbach, S. and Hancock, R.E.W. (1990) Conservation of the gene for outer membrane protein OprF in the Pseudomonaceae: Sequence of the Pseudomonas syringae oprF gene. J, Bacteriol. 31,768-775. 9 Bellido, F., Martin, N i . and Siehnel, R.J. (1992) Reevaluation in intact cells of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J. Bacteriol. 174, 5196-5203. 10 Gotoh, N., Hirokazu, W., Yoshihara, E., Nakae, T. and Nishino, T. (1989) Role of protein F in maintaining structural integrity of Pseudomonas aeruginosa outer membrane. J. Bacteriol. 171,983-990. 11 Woodruff, W.A. and Hancock, R.E.W. (1986) Expression in Escherichia coli and function of Pseudomonas aeruginosa outer membrane porin protein F. J. Bacteriol. 171, 3304-3309. 12 Counts, G.W., Schwartz, R.W., Ulness, B.K., Hamilton, D.J., Rosok, M.J., Cunningham, M.D., Tam, M.R. and Darveau, R.P. (1977) Evaluation of an immunoassay test for rapid identification of Pseudomonas aeruginosa in blood cultures. J. Clin. Microbiol. 26, 1161-1165. 13 Hancock, R.E.W., Mutharia, L.M. and Mount, E.C.A. (1985) The immunotherapeutic potential of monoclonal antibodies against P. aeruginosa protein F. Eur. J. Clin. Microbiol. 4, 224 227. 14 Gilleland, Jr., H.E., Gilleland, L.B., Hughes, E.E. and Matthews-Greer, J.M. (1992) Recombinant outer membrane protein F of Pseudomonas aeruginosa elicits antibodies that mediate opsonophagocytic killing, but not complement-mediated bacteriolysis, of various strains of P. aeruginosa. Curr. Microbiol. 24, 1-7. 15 Finnen, R.L., Martin, N.L., Siehnel, R.J. Woodruff, W.A.. Rosok, M. and Hancock, R.E.W. (1992) Analysis of structural features of the major outer membrane protein OprF from Pseudomonas aeruginosa using derivatives created by TnphoA mutagenesis and subcloning. J. Bacteriol. 174, 4977-4985. 16 Hancock, R.E.W. and Chan, L. (1988) Outer membranes of environmental isolates of Pseudomonas aeruginosa. J. Clin. Microbiol. 26, 2423-2424. 17 Mutharia, L.M. and Hancock, R.E.W. (1983) Surface localization of Pseudomonas aeruginosa outer membrane porin protein using monoclonal antibodies. Infect. Immun. 42, 1027 1033. 18 Hofstra, H., Van Tol, M.J.D. and Dankert, J. (1979) Immunofluorescent detection of the major outer membrane protein II* in Escherichia coli 026K60. FEMS Microbiol. Eett. 6, 147-150. 19 Battershill, J , Speert, D.P. and Hancock, R.E.W. (1987) Use of monoclonal antibodies against protein F of Pseudomonas aeruginosa as opsonins for phagocytosis by macrophages. Infect. lmmun. 55, 2531-2533. 20 Duchene, M., Schweizer, A., Lottspeich, F., Krauss, G., Marget, M., Vogel, K., Von Specht, B. and Domdey, It. (1988) Sequence and transcriptional start site of the P~eudomonas aeruginosa outer membrane porin protein F gene. J. Bacteriol. 170, 155-162. 21 De Mot, R., Proost, P., Van Damme, J. and Van Der Leyden, J. (1992) Homology of the root adhesin of P~'eudomonas fluomscens OE 28.3 with porin F of Pseudomonas aeruginosa and Pseudomonas syringae. Mol. Gen. Genet. 231,489-493. 22 Rutz, J.M., Abdullah, T., Singh, S.P., Kalve, V.I. and Klebba, P.E. (1991) Evolution of the ferric enterobactin receptor in Gram-negative bacteria. J. Bacteriol. 173. 5964-5974. 23 Bentley, A.T. and Klebba, P.E. (1988) Effect of lipopolysaccharide structure on reactivity of anti-porin monoclonal antibodies with the bacterial cell surface. J. Bacteriol. 170, 1063-1068. 24 Sandstron, E.G., Knapp, J.S. and Buchanan, T.B. (1982) Serology of Neisseria gonorrhoeae: W-antigen serogrouping by coagglutination and protein i serotyping by enzyme linked immunosorbent assay both detect protein I antigens. Infect. Immun. 35,229-239.