* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Visualizing Chromatin Dynamics in Cycling Cells using the
Epigenetics in stem-cell differentiation wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenomics wikipedia , lookup
Y chromosome wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
X-inactivation wikipedia , lookup
Featured Publication Note Professor Thomas Cremer and colleagues Ludwig-Maximilians-University (LMU), Munich, Germany & University of Dundee, UK Visualizing Chromatin Dynamics in Cycling Cells using the UltraVIEW VoX Whether genes can come together in specific nuclear domains for silencing/expression is a very controversially discussed topic in modern genetics. To elucidate the mechanisms that might lead to “gene kissing” events, an understanding of chromatin dynamics in the nuclei of living cells in interphase and mitosis is needed. 1 In this study, researchers at the LMU Munich tested three hypotheses first proposed by Theodor Boveri more than a hundred years ago: (1) Chromosome Territory (CT) arrangements are stably maintained during interphase, (2) Chromosome proximity patterns change profoundly during prometaphase, (3) Similar CT proximity patterns in pairs of daughter nuclei reflect symmetrical chromosomal movements during anaphase and telophase, but differ substantially from the arrangement in the mother cell nucleus. Live-cell imaging of RPE-1 cells stably expressing paGFP-HistoneH4 and mRFP-HistoneH2B was performed using the UltraVIEW VoX confocal imaging system for extremely cell-sensitive image acquisition in combination with Volocity software (version 5.2.1). A FRAP unit was used to perform photoactivation and photobleaching experiments. Photoactivation of paGFP-H4 in selected nuclear areas was performed and the resulting fluorescent chromatin patterns were traced through interphase and mitosis (figure 1). Researchers used photobleaching of H2B-mRFP fluorescence in combination with photoactivation of paGFP-H4 to test the impact of spindle position on the global transmission of chromosome proximity patterns from one cell generation to the next (figure 2). This study provided direct evidence to support Boveri’s three hypotheses and emphasizes the importance of the mechanics of mitosis for chromosome proximity patterns in interphase nuclei. Moreover, it provided new insights into the interphase flexibility of the nucleus and how it can establish long range interactions of chromatin domains in trans. 2 i Figure 1: Stable higher order chromatin arrangements in interphase nuclei Different patterns of paGFP-H4 fluorescence are stably maintained in interphase nuclei during the observation period (up to 30 h). ii iii Figure 2: Changes in chromatin proximity patterns during mitosis Upper: In roughly half of a prophase nucleus, photobleaching of mRFP-H2B (red) was performed along its short axis (i), whereas photoactivation of paGFP-H4 (green) was induced along its long axis (ii). Lower: The resulting daughter nuclei show coherent regions with mRFP fluorescence (i), but scattered patches of paGFP fluorescence (ii). Zones of overlap between red and green fluorescence are shown in (iii). Figures adapted with permission from Landes Bioscience. Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D and Cremer T (May/June 2010). 4D Chromatin dynamics in cycling cells: Theodor Boveri's hypotheses revisited (Figures 2 and 6). Nucleus; 1 (3): 1-15 © Landes Bioscience.


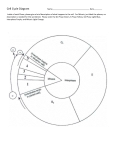







![The cell cycle multiplies cells. [1]](http://s1.studyres.com/store/data/015575697_1-eca96c262728bdb192b5eb10f1093d3e-150x150.png)