* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Unveiling the transcriptional control of the developing cardiac
Baker Heart and Diabetes Institute wikipedia , lookup
Heart failure wikipedia , lookup
Hypertrophic cardiomyopathy wikipedia , lookup
Cardiac contractility modulation wikipedia , lookup
Coronary artery disease wikipedia , lookup
Management of acute coronary syndrome wikipedia , lookup
Cardiothoracic surgery wikipedia , lookup
Cardiac surgery wikipedia , lookup
Electrocardiography wikipedia , lookup
Quantium Medical Cardiac Output wikipedia , lookup
Arrhythmogenic right ventricular dysplasia wikipedia , lookup
Cardiac arrest wikipedia , lookup
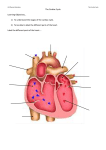
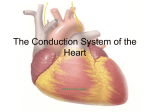
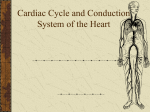
Cardiovascular Research 62 (2004) 444 – 446 www.elsevier.com/locate/cardiores Editorial Unveiling the transcriptional control of the developing cardiac conduction system Diego Franco * Department of Experimental Biology, Faculty of Health and Experimental Sciences, University of Jaén, 23071 Jaén, Spain Received 4 March 2004; accepted 10 March 2004 The development of the heart is a fascinating process. The heart starts out as a straight tube that subsequently bends to the right and forms an embryonic heart with prospective atrial and ventricular chambers [1]. Soon after, septation is initiated and the atrial and ventricular chambers are separated into left and right components, configuring a four-chambered organ with synchronous contraction. Distinct cell lineages are involved in these processes and their respective morphogenetic contributions are well described. Over the last decade, a turning point has taken place in the field of cardiovascular development since the first cardiacenriched transcription factors were discovered. Soon after, the generation of targeted mutations led to a wide range of cardiac phenotypes that has progressively enriched our understanding of cardiac (dis)morphogenesis (for a review, see Ref. [2]). Molecular interactions between different transcriptional regulators have been relatively accurately deciphered with respect to the major morphogenetic cardiac events. Interestingly, whereas there was an initial hope that a master myocardial gene would be discovered that eventually would be the key regulator of cardiac differentiation, it is now clear that the regulation of the myocardial phenotype is a complex system in which many players are involved. The advent of tissue-specific molecular markers has greatly allowed a more comprehensive understanding of cardiac development. In fact, the coupling of gene expression profile during cardiac development to other functional molecular biological techniques has permitted a more accurate understanding of cardiac (dis)morphogenesis [3]. Gene expression experiments in the developing heart already pointed out that the myocardium is rather heterogeneous (for a review, see Ref. [4]). Detailed studies using transgenic mouse models as well as endogenous gene expression profiling have demonstrated a progressive complexity of the myocardial domains as * Tel.: +34-95-3002763; fax: +34-95-3012141. E-mail address: [email protected] (D. Franco). well as their transcriptional pathways. In fact, in the early tubular heart (mouse E8.0), only homogeneous or gradient expression patterns are observed, whereas in the embryonic heart (mouse E10.5), at least five different regions can be delimited [4]. Such myocardial complexity is even more extensive in the adult heart, as recently described for the atrial chambers [5]. It is interesting to realize, nonetheless, that in the adult heart, the myocardium has been conventionally divided in two types, the fast-contracting chamber atrial and ventricular myocardium and the pace-making conducting cardiac conduction system (CCS) [6]. The existence of such a special type of myocardial cells is specifically required for the synchronous contraction of the atrial and ventricular chambers. The CCS in the adult heart can be divided into different components based on their molecular and electrophysiological characteristics [6,7]. Surprisingly, although the morphogenetic events that allow the formation of a four-chambered heart from a progenitor myocardial cell pools, including the morphological basis of the synchronous cardiac conductive properties, have been analyzed and carefully described for several decades, there is still some overt discussion about the developmental origin of the specialized CCS. It remains unknown whether the CCS is derived from the developing myocardium or else from an extracardiac origin [6,8]. It was first proposed that discrete areas of the atrioventricular canal myocardium, characterized by a glycogen-rich content, were the precursors of the atrioventricular node in the mouse [9]. Soon thereafter, a novel molecular marker, Gln2, allowed the prospective progenitor cells of the atrioventricular bundle to be traced back in humans [10]. However, the finding that some neurofilament markers were specifically expressed in the cardiac conduction system in the rabbit suggested that the neural crest cells might contribute to the development of this tissue [11]. Nowadays, a bulk of circumstantial evidence, mainly sustained by similar gene expression profiles of discrete 0008-6363/$ - see front matter D 2004 European Society of Cardiology. Published by Elsevier B.V. All rights reserved. doi:10.1016/j.cardiores.2004.03.012 D. Franco / Cardiovascular Research 62 (2004) 444–446 myocardial segments in the developing heart, clearly supports a myocardial origin of the CCS [6,12]. In addition, a number of experiments, such as cardiac neural crest ablation and retroviral cell fate experiments, have failed to obtain any direct evidence of a substantial contribution to the function and/or formation of the CCS from the cardiac neural crest cells [13]. At present, only two transcription factors (Nkx2.5 and Tbx5) have been described to play a role in cardiac conduction system function [14,15]. Nonetheless, their role during CCS development remains to be clarified. In this issue of Cardiovascular Research, Hoogaars/Tessari et al. [16] have uncovered a novel molecular marker that delineates the majority of the murine cardiac conduction system. The authors unveil the cardiac expression profile of a T-box family transcription factor, Tbx3, which is highly enriched in the fully developed CCS. Furthermore, the developmental profile of Tbx3 expression is enriched in those putative embryonic myocardial regions that have been previously proposed as early contributors of the ventricular conduction tissue [7], supporting a myocardial origin of the CCS. In addition, since Tbx3 is a transcription factor with DNA binding capabilities, these data give novel insights about the transcriptional mechanisms governing the development of the CCS. Interestingly, Hoogaars/Tessari et al. [16] provide convincing evidence of a putative mechanism by which the cardiac conduction system might escape from a working myocardium phenotype by preventing the expression of putative chamber myocardial markers such as atrial natriuretic factor. However, there are some caveats to this model that would require a refinement of our understanding on how Tbx3 acts in the developing heart. Firstly, Tbx3 is not expressed exclusively in the myocardial boundaries of the prospective cardiac conduction system components, but it also expands into the atrioventricular cushions [16]. The role of Tbx3 in the mesenchymal cushions has yet to be explored. Secondly, there are additional myocardial Tbx3-expressing areas in the fetal and adult heart that might be considered as remnants of the embryonic slow conducting areas, as suggested by the authors. However, the putative repressor role of Tbx3 in these areas becomes unclear, especially concerning the novel finding of internodal tracts between the sinoatrial and atrioventricular nodes [16]. Thirdly, and more intriguingly, Tbx3 null mutant mice lack an overt cardiac conduction system phenotype [17]. A scenario might be that a lack of Tbx3 might be compensated by Tbx2, which is also transiently expressed in the conduction system and also has a repressor DNA binding activity [18]. At present, addressing this point is compromised due to the early embryonic lethality reported in the Tbx3 mutant mice [17], which is earlier than the cardiac conduction system can be firstly traced morphologically. In this context, it is important to realize that Tbx5 is also expressed in the developing cardiac conduction system and, furthermore, that it is essential for 445 the correct morphogenesis of the conduction system [15]. Thus, from previous data and those reported in this issue by Hoogaars/Tessari et al. [16], the relevance of the T-box transcription factors in the development of the conduction system in the mouse is emerging. The generation of conditional null mutants will provide a future framework for the detailed understanding of the transcription factors of the Tbox family members that are expressed in the developing myocardium. In the same line of thinking, the emergence of new early molecular markers that can finely delineate the cardiac conduction system will be an important step forward for our understanding of the transcriptional mechanisms governing the formation of the cardiac conduction system. References [1] de Castro MP, Acosta L, Dominguez JN, Aranega A, Franco D. Molecular diversity of the developing and adult myocardium: implications for tissue targeting. Curr Drug Targets Cardiovasc Haematol Disord 2003;3:227 – 39. [2] Bruneau B. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res 2002;5:509 – 19. [3] Habets PE, Moorman AF, Christoffels VM. Regulatory modules in the developing heart. Cardiovasc Res 2003;2:246 – 63. [4] Franco D, Lamers WH, Moorman AFM. Patterns of expression in the developing myocardium: towards a morphologically integrated transcriptional model. Cardiovasc Res 1998;38:25 – 53. [5] Franco D, Campione M, Kelly R, Zammit PS, Buckingham M, Lamers WH, et al. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ Res 2000;87:984 – 91. [6] Moorman AF, de Jong F, Denyn MM, Lamers WH. Development of the cardiac conduction system. Circ Res 1998;82:629 – 44. [7] Franco D, Icardo JM. Molecular characterization of the ventricular conduction system in the developing mouse heart: topographical correlation in normal and congenitally malformed hearts. Cardiovasc Res 2001;49:417 – 29. [8] Poelmann R, Gittenberger de Groot A. A subpopulation of apoptosisprone cardiac neural crest cells targets to the venous pole: multiple functions in heart development? Dev Biol 1999;207:271 – 86. [9] Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol 1982;89:25 – 40. [10] Wessels A, Vermeulen JL, Verbeek FJ, et al. Spatial distribution of ‘‘tissue-specific’’ antigens in the developing human heart and skeletal muscle: III. An immunohistochemical analysis of the distribution of the neural tissue antigen G1N2 in the embryonic heart; implications for the development of the atrioventricular conduction system. Anat Rec 1992;232:97 – 111. [11] Gorza L, Schiaffino S, Vitadello M. Heart conduction system: a neural crest derivative? Brain Res 1988;457:360 – 6. [12] Myers DC, Fishman GI. Molecular and functional maturation of the murine cardiac conduction system. Trends Cardiovasc Med 2003;13: 289 – 95. [13] Gourdie RG, Harris BS, Bond J, et al. Development of the cardiac pacemaking and conduction system. Birth Defects Res Part C Embryo Today 2003;69:46 – 57. [14] Biben C, Weber R, Kesteven S, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res 2000;87:888 – 95. [15] Bruneau BG, Nemer G, Schmitt JP, et al. A murine model of HoltOram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001;106:709 – 21. 446 D. Franco / Cardiovascular Research 62 (2004) 444–446 [16] Hoogaars WMH, Tessari A, Moorman AFM, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovascular Research 2004;62:489 – 99. [17] Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development 2003;130: 2263 – 73. [18] Habets PE, Moorman AF, Clout DE, et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev 2002;16: 1234 – 46.