* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chapter 19 Power Point Slides
Caucasian race wikipedia , lookup
History of anthropometry wikipedia , lookup
Race (human categorization) wikipedia , lookup
Evolutionary origin of religions wikipedia , lookup
Race and health wikipedia , lookup
Race and genetics wikipedia , lookup
Human genetic clustering wikipedia , lookup
Discovery of human antiquity wikipedia , lookup
Before the Dawn (book) wikipedia , lookup
Behavioral modernity wikipedia , lookup
Human evolution wikipedia , lookup
Human genetic variation wikipedia , lookup
Human variability wikipedia , lookup
Human evolutionary genetics wikipedia , lookup
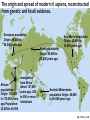
Michael Cummings Chapter 19 Population Genetics and Human Evolution David Reisman • University of South Carolina Gradients of Genetic Variation in Human Populations Prior to genomics, evolutionary biologists surveyed populations and cataloged differences in allele frequencies The distribution of a specific allele as a gradient across continents is called a cline. The gradient of an allele reflects waves of migration This data provides clues to the origin of humans— the older populations have had more generations to evolve and thus have greater diversity of alleles. http://www.genome.gov/25019968 Are There Human Races? Race—term used in the 19th century to describe phenotypic differences among populations. Studies of variations in proteins, microsatellites regions of DNA, and expressed genes show more genetic variation within populations than between populations Main Conclusion: There is no clear genetic basis for dividing our species into races Variations used in Genomic Studies Table 19-5, p. 428 Conclusions of Population Studies Based on studies from the 1990’s to the present… There is very little genetic variation in the human genome Variation in the human genome is continuously distributed Most genetic variation is widely shared, but a small amount is geographically clustered Some genetic analyses can sometimes allow the identification of an individuals continent of ancestry (see Genographic Project slide later) Australian Each circle in the center represents genetic variation within a population defined as a race. The variations overlap greatly as shown by the dark grey in the center. Few to no genetic differences belong to a single racial group. Caucasoid Human genetic variation North American ‘‘Race’’ 1 ‘‘Race’’ 2 South American African ‘‘Race’’ 3 Indian African Mainland Asian Fig. 19-9, p. 429 19.6 The Evolutionary History and Spread of Our Species (Homo sapiens) A combination of anthropology, paleontology, archaeology, and genetics is being used to reconstruct the dispersal of human populations around the globe Our evolutionary history begins with the hominoid lineage about 25 million years ago Hominoid – superfamily of primates, including apes and humans Variations on Rudolph Zallinger’s March of Progress Images from http://christpantokrator.blogspot.com/2011/06/human-evolution-diagrams.html Evolution is not a linear morphing of one type of organism into another – a common misunderstanding http://pewresearch.org/pubs/1105/darwin-debate-religion-evolution http://tolweb.org/onlinecontributors/app Rather, evolution is branching from a shared ancestor Genome-based evolutionary relationships among hominoids 7 m.y.a. chimps and humans had a common ancestor Fig. 19-10, p. 430 Early humans emerged ~5 million years ago After human line split from the chimps, three different species groups appeared. Collectively known as hominins • Australopithecines • Paranthropus • Homo – our ancestral group Homo floresiensis Homo rudolfensis Australopithecus anamensis Homo sapiens Homo habilis Australopithecus africanus Australopithecus afarensis Homo erectus Australopithecus garhi Paranthropus aethiopicus Homo neanderthalensis Paranthropus robustus Paranthropus boisei 4 3 2 1 Present Time (millions of years ago) Estimates of the dates of origin and extinction of the three main groups of hominins (green, blue, and orange). The australopithecines split into two groups about 2.5 to 2.7 million years ago. Fig. 19-11, p. 430 Two Theories Differ on How and Where Homo sapiens Originated Hypothesis 1: Modern Homo sapiens arose once, in one place, from its ancestral species. Members of this species then migrated from there to all parts of the globe. (Out-of-Africa hypothesis) Hypothesis 2: Modern Homo sapiens arose in a number of different locations from similar ancestral populations at roughly the same time. The hypothesis assumes that although the populations were in different locations, they did interbreed and exchange genes. (Multiregional hypothesis) Genetic evidence supports model #1. Humans Have Spread Across the World Available evidence suggests that • H. sapiens emigrated from Africa about 137,000 years ago • H. sapiens spread through Southeast Asia and Australia 40,000 to 60,000 years ago • H. sapiens replaced Neanderthals in Europe 40,000 to 50,000 years ago • North America and South America were populated in waves 15,000 to 30,000 years ago The origin and spread of modern H. sapiens, reconstructed from genetic and fossil evidence. European population Origin: 40,000 to 50,000 years ago Asian population Origin: 50,000 to 70,000 years ago Immigration from Africa African About 137,000 populations years ago; 200 Origin: 130,000 to 500 or more to 170,000 years individuals ago Population: 23,000 to 45,000 New World population Origin: 20,000 to 30,000 years ago Australo-Melanesian population Origin: 40,000 to 60,000 years ago Fig. 19-13, p. 432 *All human populations are derived from African populations **Colors correspond to major continental regions. Fig. 19-12, p. 431 19.7 Genomics and Human Evolution Although separated for about 7 million years, analysis of human and chimp genomes shows many similarities and subtle differences • The DNA sequences are 98.8% identical • Variations due to insertions, deletions and duplications differ, ultimately change gene dosage • There is a 1% difference in coding sequence of genes • Phenotypic differences cannot be explained by differences in coding sequences • The important differences may involve gene regulation and genes that control body structure Neanderthals are not Closely Related to Humans H. neanderthalensis lived in the Middle East, Asia, and Europe 300,000 to 30,000 years ago. Analysis of DNA recovered from Neanderthal remains clearly show that humans did not descend from them. (Neanderthal genome sequenced in 2010. Some interbreeding did occur most likely in the Middle East before humans expanded into Europe and Asia 1-4% of genes carried by non-africans are from Neanderthals –440,000 to 270,000 y. a. Split of ancestral human and Neanderthal populations –706,000 y. a. Coalescence of human and Neanderthal reference sequences –41,000 y. a. Earliest modern humans in Europe –195,000 y. a. Earliest known anatomically modern humans –28,000 y. a. Most recent known Neanderthal remains Modern human Neanderthal Genomic data Fossil data Genomic and fossil evidence has been used to estimate the time of divergence of human and Neanderthal lines relative to landmark events evolution. Genomic analyses trace evolution back much farther than fossils can. Fig. 19-15, p. 434 DNA as a Molecular Clock The rate of mutation between two DNA sequences can be used as a clock to provide a relative measure of time since divergence from a common ancestor Assumes that mutation rate is constant Can be calibrated by comparison to the fossil record The Genetic Revolution: Tracing Ancient Migrations How can we map out events that occurred thousands of years ago? • The answers are written in the genomes of present day populations • Genetic markers on the Y chromosome are passed from father to son • Markers in mitochondrial DNA are passed from mother to all offspring • These markers do not undergo recombination in meiosis—individuals carry these markers to new locations as the migrate • The Genographic Project: https://genographic.nationalgeographic.com/genographi c/index.html An Important Gene in Language Development The gene is called FOXP2 It is present in chimpanzees, modern humans, and Neanderthals Genes and pathways controlled by FOXP2 differ among these groups http://www.physorg.com/news188139245.html With the help of a little singing bird, Penn State physicists are gaining insight into how the human brain functions, which may lead to a better understanding of complex vocal behavior, human speech production and ultimately, speech disorders and related diseases.