* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download lecture 47 slides no animations
Fatty acid metabolism wikipedia , lookup
Magnesium transporter wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Interactome wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
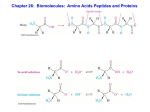
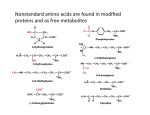
Lecture 47: Structure II -- Proteins Today’s Outline • The monomers: amino acids – Side chain characteristics – Acid-base equilibria and pKa • Peptide backbone – Peptide bond formation – Rotational degrees of freedom – Common folding motifs • Side chain interactions in folding • (Time permitting) Levinthal’s paradox This is not intended as a Fischer projection! First AA in most proteins Bulkiest isoleucine Isomers Smallest Covalent bond between side chain and These three are the most commonly phosphorylated residues in eukaryotes Umami flavor of MSG Neurotransmitter As appropriate a time as any: pKa As appropriate a time as any: pKa Rearranging, we get: When pH >> pKa, [A-] >> [HA] When pH << pKa, [A-] << [HA] When pH = pKa, [A-] = [HA] Strong acids have low pKas: • HBr: -10 • HCl: -7 Weaker acids have low-ish pKas: • H2PO4: 2 • HF: 3 • D/E: 4 Umami flavor of MSG Neurotransmitter pKa = 3.9 pKa = 4.1 pKa = 10.5 pKa = 12.5 pKa = 6.0 Peptide bonds link amino acids A protein’s folded shape can be roughly described by the path traced by its peptide backbone (ignoring sidechains) Peptide bonds have limited rotational freedom The peptide bond is planar: The peptide bond has two conformations …but cis is less likely due to steric clash Most backbone rotation occurs via phi (f) and psi (y) torsion angles Some combinations of torsion angles are much more likely than others Ramachandran plot: shows frequency of (f,y) observed for residues in folded proteins Ball-and-stick models provide some insight on infrequency of f >0 Glycine does adopt positive f values Less steric hindrance because side chain (green) is very small Proline’s f value is (somewhat) fixed by its side chain’s bond to the backbone This side chain can clash sterically with the preceding amino acid, so the“preproline” Ramachandran plot is also unique Pre-proline Ramachandran plot Another trend for consecutive amino acids: correlation in position Amino acid n Amino acid n+1 What do we get if we repeat the same torsion angles many times in a row? PyMOL interactive PyMOL interactive These amino acids participate in the more common, right-handed form of helices PyMOL interactive Other right-handed helices besides alpha helices have similar torsion angles a helix 310 helix p helix PyMOL interactive PyMOL interactive These amino acids are part of beta sheets PyMOL non-interactive These amino acids participate in the less common, left-handed forms of helices Most of the residues in most folded proteins participate in one of these motifs Peptide backbone hydrogen bonding is the most common motif in folding • The pattern of backbone hydrogen bonding is referred to as a protein’s secondary structure • Some side chains inhibit formation of certain secondary structures (e.g. proline/a helices) • With those exceptions, secondary structures are not dependent on amino acid sequence Does hydrogen bonding in secondary structures drive protein folding? What interactions between side chains drive sequence-specific protein folding? Hydrophobic effect Steric clash What interactions between side chains drive sequence-specific protein folding? These forces determine the relative orientation of a protein’s secondary structures, i.e., the protein’s tertiary structure The same forces that drive tertiary structure formation can also hold two or more proteins together Hydrophobic effect and vdW forces drive leucine zipper folding Covalent disulfide bonds can form between cysteines Reduction potentials: Cysteines: -222 mV NADH: -320 mV Formal collaboration policy This course encourages collaboration, which is a key to learning and the progress of science. You may talk together about approaches to problems and you may work together to perform calculations or write code. But any formal assignments that you are given must be written, rather than copied, by you and you only, with the exception of computer code, where the code may be written communally, but all code must be well commented, and all comments in graded code must be written by you, on your own. Since the goal of assignments is to help you and us assess your level of understanding of the material, we require that you understand both the conceptual structure and the details of each individual step of any answer you submit and we may ask you to answer questions, orally, about an assignment, to demonstrate that you have achieved this level of understanding.