Framework for Teachable Unit
... 2) reduces affinity for other nucleosomes, reducing tightness of the second order structure Methylation 1) No change in charge but methylation recruits silencing or repressive proteins to the site ...
... 2) reduces affinity for other nucleosomes, reducing tightness of the second order structure Methylation 1) No change in charge but methylation recruits silencing or repressive proteins to the site ...
Mitosis Vocabulary Review
... b. chromatin d. centromere _____ 12. In eurkaryotes, a structural unit made up of DNA wound around a center of histone proteins is called a a. chromatid. c. centrosome. b. nucleosome. d. looped domain. _____ 13. The structure that directs chromosome movement during mitosis and aids in the formation ...
... b. chromatin d. centromere _____ 12. In eurkaryotes, a structural unit made up of DNA wound around a center of histone proteins is called a a. chromatid. c. centrosome. b. nucleosome. d. looped domain. _____ 13. The structure that directs chromosome movement during mitosis and aids in the formation ...
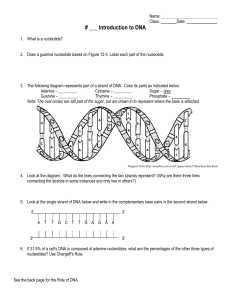
Cytosine – ______ Sugar
... Note: The oval circles are still part of the sugar, but are drawn in to represent where the base is attached. ...
... Note: The oval circles are still part of the sugar, but are drawn in to represent where the base is attached. ...
MCAS BIOLOGY REVIEW GENETICS AND EVOLUTION
... Describe the basic structure (double helix, sugar/phosphate backbone, linked by complementary nucleotide pairs) of DNA, and describe its function in genetic inheritance. What is DNA: http://www.statedclearly.com/what-is-dna/ ...
... Describe the basic structure (double helix, sugar/phosphate backbone, linked by complementary nucleotide pairs) of DNA, and describe its function in genetic inheritance. What is DNA: http://www.statedclearly.com/what-is-dna/ ...
Chapter 1 Answers
... would this be the case? The DNA is not visible during interphase because during this phase, portions are being used in transcription of mRNA for protein synthesis. DNA is in an “unraveled” configuration that will allow segments to be “unzipped” and transcribed. During interphase, the cell is carryin ...
... would this be the case? The DNA is not visible during interphase because during this phase, portions are being used in transcription of mRNA for protein synthesis. DNA is in an “unraveled” configuration that will allow segments to be “unzipped” and transcribed. During interphase, the cell is carryin ...
Genetic Engineering Short Notes
... 1. Genetic engineering- remaking genes for practical purposes 2. Recombinant DNA- DNA made from two or more different organisms 3. Restriction enzyme- enzymes that recognize short specific DNA sequences and that cut the DNA there 4. Plasmid- small, circular DNA molecules that can replicate independa ...
... 1. Genetic engineering- remaking genes for practical purposes 2. Recombinant DNA- DNA made from two or more different organisms 3. Restriction enzyme- enzymes that recognize short specific DNA sequences and that cut the DNA there 4. Plasmid- small, circular DNA molecules that can replicate independa ...
REPLICATION, TRANSCRIPTION, TRANSLATION TAKS
... 26 If a cat has 38 chromosomes in each of its body cells, how many chromosomes will be in each daughter cell after mitosis? F 11 G 19 H* 38 J 76 APRIL 2004 – 11: ...
... 26 If a cat has 38 chromosomes in each of its body cells, how many chromosomes will be in each daughter cell after mitosis? F 11 G 19 H* 38 J 76 APRIL 2004 – 11: ...
New Ligands of CRABP2 Suggest a Role for this Protein in
... Retinoic acid (RA) regulates transcription of a series of genes involved in cell proliferation, differentiation and apoptosis by binding to the RA receptor (RAR) and retinoid X receptor (RXR) heterodimers. The cellular retinoic acid-binding protein 2 (CRABP2) is involved in the transport of RA from ...
... Retinoic acid (RA) regulates transcription of a series of genes involved in cell proliferation, differentiation and apoptosis by binding to the RA receptor (RAR) and retinoid X receptor (RXR) heterodimers. The cellular retinoic acid-binding protein 2 (CRABP2) is involved in the transport of RA from ...
01 - edl.io
... b. chromatin d. centromere _____ 12. In eurkaryotes, a structural unit made up of DNA wound around a center of histone proteins is called a a. chromatid. c. centrosome. b. nucleosome. d. looped domain. _____ 13. The structure that directs chromosome movement during mitosis and aids in the formation ...
... b. chromatin d. centromere _____ 12. In eurkaryotes, a structural unit made up of DNA wound around a center of histone proteins is called a a. chromatid. c. centrosome. b. nucleosome. d. looped domain. _____ 13. The structure that directs chromosome movement during mitosis and aids in the formation ...
Procaryotic chromosome
... 2. Eukaryotic chromatin: Histones (octamer)+146bp DNA > Nucleosome core + H1 >chromatosome + Linker DNA (10--55200+) > beads on string > 30nm fiber > fiber loop (to 100bp) +nuclear matrix > chromosome 3. Jargons: centromere, kinetochore, telomere, hetero or euchromatin, CpG island and methylation 4. ...
... 2. Eukaryotic chromatin: Histones (octamer)+146bp DNA > Nucleosome core + H1 >chromatosome + Linker DNA (10--55200+) > beads on string > 30nm fiber > fiber loop (to 100bp) +nuclear matrix > chromosome 3. Jargons: centromere, kinetochore, telomere, hetero or euchromatin, CpG island and methylation 4. ...
Name: Genetics Study Guide
... What does codominance mean in genetics? How is it different from Incomplete dominance? Know the difference between a hybrid and a purebred. In what decade was the DNA structure discovered? Who discovered the structure of DNA? What is the scientific name of the DNA structure? Which is the correct ord ...
... What does codominance mean in genetics? How is it different from Incomplete dominance? Know the difference between a hybrid and a purebred. In what decade was the DNA structure discovered? Who discovered the structure of DNA? What is the scientific name of the DNA structure? Which is the correct ord ...
a10c Biotechnology
... 2. What is a restriction enzyme, and what does it catalyze? How do restriction enzymes differ in what they cleave? What do they "look for"? Name an example of a restriction enzyme. 3. Describe the steps of cloning (transferring a gene to bacteria for purposes of "growing" DNA or protein). What enzym ...
... 2. What is a restriction enzyme, and what does it catalyze? How do restriction enzymes differ in what they cleave? What do they "look for"? Name an example of a restriction enzyme. 3. Describe the steps of cloning (transferring a gene to bacteria for purposes of "growing" DNA or protein). What enzym ...
3-10
... Subject: The structure and replication of DNA. Reading in ‘An introduction to genetic analysis’ (Griffiths et al., 7th edition) Chapter 8: The structure and replication of DNA. ________________________________________________________________________ Key concepts and keywords: DNA: the genetic materi ...
... Subject: The structure and replication of DNA. Reading in ‘An introduction to genetic analysis’ (Griffiths et al., 7th edition) Chapter 8: The structure and replication of DNA. ________________________________________________________________________ Key concepts and keywords: DNA: the genetic materi ...
This examination paper consists of 4 pages
... can be as short as 100 bp occur in prokaryotes and in eukaryotes transpose conservatively code for a transposase enzyme ...
... can be as short as 100 bp occur in prokaryotes and in eukaryotes transpose conservatively code for a transposase enzyme ...
E:Med - uni-freiburg.de
... Martin Vingron’s group • Sequence alignment • Microarray gene analysis • Gene regulation and evolution: – (combinatorial) TF DNA binding prediction – Histone modification gene expression – Factors affecting mutation rates ...
... Martin Vingron’s group • Sequence alignment • Microarray gene analysis • Gene regulation and evolution: – (combinatorial) TF DNA binding prediction – Histone modification gene expression – Factors affecting mutation rates ...
Cell Metabolism
... Thousands of molecules in each cell continually reacting with each other to maintain cell function DNA directs cell metabolism by instructing the cell to make proteins Do this now; hand it in Read: Whale book pg 1801 Answer these Q’s with complete thoughts & sentences: • What determines the shap ...
... Thousands of molecules in each cell continually reacting with each other to maintain cell function DNA directs cell metabolism by instructing the cell to make proteins Do this now; hand it in Read: Whale book pg 1801 Answer these Q’s with complete thoughts & sentences: • What determines the shap ...
Nucleosome

A nucleosome is a basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound in sequence around eight histone protein cores. This structure is often compared to thread wrapped around a spool.Nucleosomes form the fundamental repeating units of eukaryotic chromatin, which is used to pack the large eukaryotic genomes into the nucleus while still ensuring appropriate access to it (in mammalian cells approximately 2 m of linear DNA have to be packed into a nucleus of roughly 10 µm diameter). Nucleosomes are folded through a series of successively higher order structures to eventually form a chromosome; this both compacts DNA and creates an added layer of regulatory control, which ensures correct gene expression. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones.Nucleosomes were observed as particles in the electron microscope by Don and Ada Olins and their existence and structure (as histone octamers surrounded by approximately 200 base pairs of DNA) were proposed by Roger Kornberg. The role of the nucleosome as a general gene repressor was demonstrated by Lorch et al. in vitro and by Han and Grunstein in vivo.The nucleosome core particle consists of approximately 147 base pairs of DNA wrapped in 1.67 left-handed superhelical turns around a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4. Core particles are connected by stretches of ""linker DNA"", which can be up to about 80 bp long. Technically, a nucleosome is defined as the core particle plus one of these linker regions; however the word is often synonymous with the core particle. Genome-wide nucleosome positioning maps are now available for many model organisms including mouse liver and brain.Linker histones such as H1 and its isoforms are involved in chromatin compaction and sit at the base of the nucleosome near the DNA entry and exit binding to the linker region of the DNA. Non-condensed nucleosomes without the linker histone resemble ""beads on a string of DNA"" under an electron microscope.In contrast to most eukaryotic cells, mature sperm cells largely use protamines to package their genomic DNA, most likely to achieve an even higher packaging ratio. Histone equivalents and a simplified chromatin structure have also been found in Archea, suggesting that eukaryotes are not the only organisms that use nucleosomes.