Chapter 18 Gene Regulation
... differential gene expression, the expression of different genes by cells with the same genome • Abnormalities in gene expression can lead to diseases including cancer • Gene expression is regulated at many stages ...
... differential gene expression, the expression of different genes by cells with the same genome • Abnormalities in gene expression can lead to diseases including cancer • Gene expression is regulated at many stages ...
7.3 Protein Synthesis
... P-site and bonds by base pairing its anti-codon with the mRNA start codon (what is the start codon?) • Elongation – The second tRNA then comes into A-site and bonds to codon of mRNA – The two amino acids joined with peptide bond • Termination – ribosome continues reading mRNA until a STOP codon is r ...
... P-site and bonds by base pairing its anti-codon with the mRNA start codon (what is the start codon?) • Elongation – The second tRNA then comes into A-site and bonds to codon of mRNA – The two amino acids joined with peptide bond • Termination – ribosome continues reading mRNA until a STOP codon is r ...
Protein Synthesis 2013
... P-site and bonds by base pairing its anti-codon with the mRNA start codon (what is the start codon?) • Elongation – The second tRNA then comes into A-site and bonds to codon of mRNA – The two amino acids joined with peptide bond • Termination – ribosome continues reading mRNA until a STOP codon is r ...
... P-site and bonds by base pairing its anti-codon with the mRNA start codon (what is the start codon?) • Elongation – The second tRNA then comes into A-site and bonds to codon of mRNA – The two amino acids joined with peptide bond • Termination – ribosome continues reading mRNA until a STOP codon is r ...
CHEM642-07 Powerpoint
... must be traversed in a 3' to 5' direction. Thus, the direction of RNA polymerase movement determines which of the two DNA strands is to serve as a template for the synthesis of RNA, as shown in (A) and (B). Polymerase direction is, in turn, determined by the orientation of the promoter sequence, the ...
... must be traversed in a 3' to 5' direction. Thus, the direction of RNA polymerase movement determines which of the two DNA strands is to serve as a template for the synthesis of RNA, as shown in (A) and (B). Polymerase direction is, in turn, determined by the orientation of the promoter sequence, the ...
WELCOME TO BIOLOGY 2002 - University of Indianapolis
... Translation occurs in three steps: • Elongation: amino acids are joined together and the ribosome moves to the next codon. • New tRNAs enters the A site of the ribosome • A peptide bond forms between the polypeptide on the tRNA in the P site and the amino acid in the A site, which transfers the poly ...
... Translation occurs in three steps: • Elongation: amino acids are joined together and the ribosome moves to the next codon. • New tRNAs enters the A site of the ribosome • A peptide bond forms between the polypeptide on the tRNA in the P site and the amino acid in the A site, which transfers the poly ...
CHAPTER 17 - HCC Learning Web
... • During the elongation stage, amino acids are added one by one to the preceding amino acid at the C-terminus of the growing chain • Each addition involves proteins called elongation factors and occurs in three steps: codon recognition, peptide bond formation, and ...
... • During the elongation stage, amino acids are added one by one to the preceding amino acid at the C-terminus of the growing chain • Each addition involves proteins called elongation factors and occurs in three steps: codon recognition, peptide bond formation, and ...
Lecture ten
... in the same organism – the genomes are identical from cell to cell so why do different cells express different genes/proteins?? differences result from differential gene expression = the expression of different genes by cells with the same genome several steps along the replication/transcription/tra ...
... in the same organism – the genomes are identical from cell to cell so why do different cells express different genes/proteins?? differences result from differential gene expression = the expression of different genes by cells with the same genome several steps along the replication/transcription/tra ...
Dear Jennifer - Ms. V Biology
... Include in your explanation the words amino acid, anti-codon, codon, cytoplasm, DNA, mRNA, nucleotide, nucleus, ribosome, RNA polymerase, tRNA, transcription, and translation. (Hint: You can use the answer to question 2 on page 5 for the beginning of the answer to this question.) ...
... Include in your explanation the words amino acid, anti-codon, codon, cytoplasm, DNA, mRNA, nucleotide, nucleus, ribosome, RNA polymerase, tRNA, transcription, and translation. (Hint: You can use the answer to question 2 on page 5 for the beginning of the answer to this question.) ...
Gene expression Most genes are not expressed at a particular time
... Transcription factor binding sites Transcription factors and their binding sites come in several varieties: – Promoters are required for RNA polymerase to bind and begin transcription. The promoter binding site is often a TATAAT ~10bp upstream of the start of transcription. – Activators are proteins ...
... Transcription factor binding sites Transcription factors and their binding sites come in several varieties: – Promoters are required for RNA polymerase to bind and begin transcription. The promoter binding site is often a TATAAT ~10bp upstream of the start of transcription. – Activators are proteins ...
Reliable transfer of transcriptional gene regulatory networks
... the original and the transferred database content. Figure 2a exemplarily shows a network visualization of the PcaR regulon as known from CG compared to the transferred one of CE. It is obvious that all of the 11 target genes including the regulator itself are orthologous between CG and CE. Since the ...
... the original and the transferred database content. Figure 2a exemplarily shows a network visualization of the PcaR regulon as known from CG compared to the transferred one of CE. It is obvious that all of the 11 target genes including the regulator itself are orthologous between CG and CE. Since the ...
triplex-forming oligonucleotide (TFO)
... • Hydroxyurea – However, many patients cannot achieve increased HbF with these treatments! – With hydroxyurea treatment, for example, only about 60% of patients were found to ...
... • Hydroxyurea – However, many patients cannot achieve increased HbF with these treatments! – With hydroxyurea treatment, for example, only about 60% of patients were found to ...
Slide 1
... Protein coding gene - A DNA sequence coding for a single polypeptide Gene expression – mechanism by which hereditary factors are coded for and expressed Genes control inherited variation via: DNA, RNA and protein *Transcription – transfer of genetic information from DNA via synthesis of RNA *Transla ...
... Protein coding gene - A DNA sequence coding for a single polypeptide Gene expression – mechanism by which hereditary factors are coded for and expressed Genes control inherited variation via: DNA, RNA and protein *Transcription – transfer of genetic information from DNA via synthesis of RNA *Transla ...
Transcription - OpenStax CNX
... • Explain the main steps of transcription • Describe how eukaryotic mRNA is processed In both prokaryotes and eukaryotes, the second function of DNA (the rst was replication) is to provide the information needed to construct the proteins necessary so that the cell can perform all of its functions. ...
... • Explain the main steps of transcription • Describe how eukaryotic mRNA is processed In both prokaryotes and eukaryotes, the second function of DNA (the rst was replication) is to provide the information needed to construct the proteins necessary so that the cell can perform all of its functions. ...
Oc - TUM
... specificity of gene regulation depends on specific protein-DNA interactions mediated by the chemical interactions between aa side chains and chemical groups of DNA bases ...
... specificity of gene regulation depends on specific protein-DNA interactions mediated by the chemical interactions between aa side chains and chemical groups of DNA bases ...
manual HiScribe T7 In Vitro Transcription Kit E2030
... ~1.5 mg per ml of transcription reaction should be easily obtainable using the HiScribe T7 In Vitro Transcription Kit. A 40 µl pilot reaction should be carried out initially to test the quality of DNA template and transcription reagents. Reactions can then be scaled up accordingly as required by the ...
... ~1.5 mg per ml of transcription reaction should be easily obtainable using the HiScribe T7 In Vitro Transcription Kit. A 40 µl pilot reaction should be carried out initially to test the quality of DNA template and transcription reagents. Reactions can then be scaled up accordingly as required by the ...
Chapter 1 Notes
... - mutations that affect eye color in Drosophila block pigment synthesis at a specific step by preventing production of the enzyme that catalyzes that step -b/c each mutant was defective in a single gene, the function of a gene is to dictate the production of an enzyme ...
... - mutations that affect eye color in Drosophila block pigment synthesis at a specific step by preventing production of the enzyme that catalyzes that step -b/c each mutant was defective in a single gene, the function of a gene is to dictate the production of an enzyme ...
on-chip
... • The implication is that ISWI chromatin remodelers can organize chromatin into a more repressive configuration, OR a more active one • Since yeast has two ISWI ATPases with different effects on chromatin in vitro, they may have evolved contrasting activities ...
... • The implication is that ISWI chromatin remodelers can organize chromatin into a more repressive configuration, OR a more active one • Since yeast has two ISWI ATPases with different effects on chromatin in vitro, they may have evolved contrasting activities ...
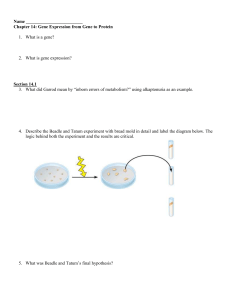
Chapter 14 Guided Reading
... 3. What did Garrod mean by “inborn errors of metabolism?” using alkaptonuria as an example. ...
... 3. What did Garrod mean by “inborn errors of metabolism?” using alkaptonuria as an example. ...
Regulation of gene expression: Prokaryotic
... • In bacteria, mRNA is polycistronic; in eukaryotes, mRNA is usually monocistronic. – Polycistronic: one mRNA codes for more than one polypeptide – moncistronic: one mRNA codes for only one polypeptide • 3 RNA polymerases in euk., 1 in prok. Binding of Basal Transcription Factors required for euk. R ...
... • In bacteria, mRNA is polycistronic; in eukaryotes, mRNA is usually monocistronic. – Polycistronic: one mRNA codes for more than one polypeptide – moncistronic: one mRNA codes for only one polypeptide • 3 RNA polymerases in euk., 1 in prok. Binding of Basal Transcription Factors required for euk. R ...
Transcriptional activation by the human herpesvirus-8
... important in mediating transactivation by certain transcription factors such as IRF-1, transcriptional repression and activation by vIRF act, at least in part, through distinct mechanisms. A vIRF truncation mutant lacking the N-terminal 151 amino acids (vIRF 152–449 ; NTvIRF) retains wild-type abili ...
... important in mediating transactivation by certain transcription factors such as IRF-1, transcriptional repression and activation by vIRF act, at least in part, through distinct mechanisms. A vIRF truncation mutant lacking the N-terminal 151 amino acids (vIRF 152–449 ; NTvIRF) retains wild-type abili ...
Bio-261-chapter-7
... The exception to this is that uracil is used for nucleotide sequencing of RNA molecules rather than thymine. ...
... The exception to this is that uracil is used for nucleotide sequencing of RNA molecules rather than thymine. ...
C - TeacherWeb
... The exception to this is that uracil is used for nucleotide sequencing of RNA molecules rather than thymine. ...
... The exception to this is that uracil is used for nucleotide sequencing of RNA molecules rather than thymine. ...
Recommendations for Riboprobe Synthesis
... and delivery of specimens to the Core. Alternatively, the Core provides archival tissues and embryos from ICR outbred mice in paraffin that have been previously fixed for RNA integrity. - Preparation of Tissues - The Core is responsible for paraffin processing, embedding, and preparing histologic se ...
... and delivery of specimens to the Core. Alternatively, the Core provides archival tissues and embryos from ICR outbred mice in paraffin that have been previously fixed for RNA integrity. - Preparation of Tissues - The Core is responsible for paraffin processing, embedding, and preparing histologic se ...
Chromatin Structure and Its Effects on Transcription
... DNA/core histones when contrasted with: – Uncatalyzed DNA exposure in nucleosomes – Simple nucleosome sliding along a DNA stretch ...
... DNA/core histones when contrasted with: – Uncatalyzed DNA exposure in nucleosomes – Simple nucleosome sliding along a DNA stretch ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.