I. Comparing genome sequences
... • Genomes become more dissimilar with greater phylogenetic distance ...
... • Genomes become more dissimilar with greater phylogenetic distance ...
I. Comparing genome sequences
... • Genomes become more dissimilar with greater phylogenetic distance ...
... • Genomes become more dissimilar with greater phylogenetic distance ...
Phylogenetic relationships among iguanian lizards using alternative
... mixture model that employs a reversible-jump algorithm to estimate the number of rate matrices that best explains the data. This method chooses the appropriate number of independent rate matrices using Bayes factors during the MCMC procedure. By default, BayesPhylogenies assigns uniform priors on a ...
... mixture model that employs a reversible-jump algorithm to estimate the number of rate matrices that best explains the data. This method chooses the appropriate number of independent rate matrices using Bayes factors during the MCMC procedure. By default, BayesPhylogenies assigns uniform priors on a ...
genomic diversity and differentiation
... π, average number of differences among sequences (what is it below?) ηi, folded site pattern: how many segregating sites appear i times? ...
... π, average number of differences among sequences (what is it below?) ηi, folded site pattern: how many segregating sites appear i times? ...
Proposal to change linear sequence of orders to place Galliformes
... Synopsis: An accumulating body of independent evidence indicates that the (1) the Galliformes and Anseriformes are sister taxa, and (2) they represent the earliest branch in the living class Aves after the palaeognaths. These two orders have traditionally been separated by the Falconiformes and plac ...
... Synopsis: An accumulating body of independent evidence indicates that the (1) the Galliformes and Anseriformes are sister taxa, and (2) they represent the earliest branch in the living class Aves after the palaeognaths. These two orders have traditionally been separated by the Falconiformes and plac ...
Supplemental Data
... Figure S7 Phenotypes of double mutant combinations between major and minor function genes. (A) Wild-type, (B) irx10 irx14-L, (C) irx14 irx9-L and (D) irx14 irx10-L, (E) irx9, (F) irx10-L, (G) irx9 irx14-L and (H) irx9 irx10 5 week old soil grown plants. (I) From left to right are wt, irx10-L f8h, ir ...
... Figure S7 Phenotypes of double mutant combinations between major and minor function genes. (A) Wild-type, (B) irx10 irx14-L, (C) irx14 irx9-L and (D) irx14 irx10-L, (E) irx9, (F) irx10-L, (G) irx9 irx14-L and (H) irx9 irx10 5 week old soil grown plants. (I) From left to right are wt, irx10-L f8h, ir ...
Multiple Sequence Alignment
... • PAM30 for closely related pairs; PAM120; PAM250 for more distant ...
... • PAM30 for closely related pairs; PAM120; PAM250 for more distant ...
Ch268thed
... Distinguishes between shared primitive and shared derived characteristics Closely related to ingroup ...
... Distinguishes between shared primitive and shared derived characteristics Closely related to ingroup ...
Presentazione di PowerPoint
... Usually, members of genes families are sharing a high homology in their sequence, and, when they are functionally active, they perform very similar biological functions: they are called paralog genes or simply paralogs. Endogenous mechanisms represent quantitatively the most important process that l ...
... Usually, members of genes families are sharing a high homology in their sequence, and, when they are functionally active, they perform very similar biological functions: they are called paralog genes or simply paralogs. Endogenous mechanisms represent quantitatively the most important process that l ...
Species Tree and Most Likely Gene Tree
... are short, frequently there isn’t enough information about that specific branching because very few mutations happen along it. In those cases, anomalous gene trees aren’t even an issue. So in actual sequence analysis, anomalous gene trees may only come up when these short internal branches have high ...
... are short, frequently there isn’t enough information about that specific branching because very few mutations happen along it. In those cases, anomalous gene trees aren’t even an issue. So in actual sequence analysis, anomalous gene trees may only come up when these short internal branches have high ...
Sequence Weights - Semantic Scholar
... Well-formulated as an optimization problem. Independent of sequence order. Uses all information. Tree may be rooted anywhere, allowing outgroups to contribute. Possible disadvantages: Leaves farther from the root are downweighted. Assumes an evolutionary tree relating the sequence. Major disadvantag ...
... Well-formulated as an optimization problem. Independent of sequence order. Uses all information. Tree may be rooted anywhere, allowing outgroups to contribute. Possible disadvantages: Leaves farther from the root are downweighted. Assumes an evolutionary tree relating the sequence. Major disadvantag ...
LS50B Concept questions: end of section 6: Solutions
... • A polyphyletic group is a group of organisms that do not have their most recent common ancestor in common (i.e. at least one of them has a sister clade that’s not in the group). • A paraphyletic group includes a given common ancestor and some, but not all, of its descendants. 3. Why should we care ...
... • A polyphyletic group is a group of organisms that do not have their most recent common ancestor in common (i.e. at least one of them has a sister clade that’s not in the group). • A paraphyletic group includes a given common ancestor and some, but not all, of its descendants. 3. Why should we care ...
manual
... the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNE ...
... the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNE ...
Modelling_evolution - the Department of Statistics
... with the 64 x 64 matrix of states that are the codons (Goldman and Yang ...
... with the 64 x 64 matrix of states that are the codons (Goldman and Yang ...
Language Trees
... which describes the way several languages are related. Latin is the ancestor, and the “leaves” are the contemporary languages. ...
... which describes the way several languages are related. Latin is the ancestor, and the “leaves” are the contemporary languages. ...
File
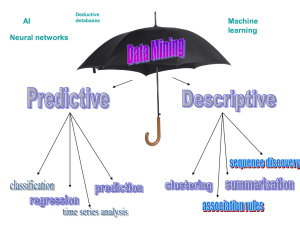
... The main idea of decision tree construction tree is to evaluate different attributes and different partitioning conditions, and pick the attributes and partitioning condition that results in the maximum information gain ratio. The same procedure works recursively on each of the sets resulting from t ...
... The main idea of decision tree construction tree is to evaluate different attributes and different partitioning conditions, and pick the attributes and partitioning condition that results in the maximum information gain ratio. The same procedure works recursively on each of the sets resulting from t ...
tutorial_em - NYU Computer Science
... Inject noise in covariance matrix to prevent blowup Single point gives infinite likelihood Number of components Open problem Minimum description length Bayesian approach ...
... Inject noise in covariance matrix to prevent blowup Single point gives infinite likelihood Number of components Open problem Minimum description length Bayesian approach ...