Phylogenetic Relationships Among Ascomycetes: Evidence from an
... Sequence Alignment and Phylogenetic Analyses The predicted amino acid sequences of RPB2 between regions 3 and 11 of 26 fungi were aligned to RPB2 sequences of S. cerevisiae and S. pombe using CLUSTAL V (Higgins, Bleasby, and Fuchs 1992), with subsequent manual adjustment. The resultant alignment con ...
... Sequence Alignment and Phylogenetic Analyses The predicted amino acid sequences of RPB2 between regions 3 and 11 of 26 fungi were aligned to RPB2 sequences of S. cerevisiae and S. pombe using CLUSTAL V (Higgins, Bleasby, and Fuchs 1992), with subsequent manual adjustment. The resultant alignment con ...
knn - MSU CSE
... • A small neighborhood large variance unreliable estimation • A large neighborhood large bias inaccurate estimation ...
... • A small neighborhood large variance unreliable estimation • A large neighborhood large bias inaccurate estimation ...
Investigating Polar Bear and Giant Panda Ancestry
... This activity will allow you to use the tools used by geneticists and evolutionists located and maintained by the National Center for Biotechnology Information. These tools will allow you to compare the isolated and sequenced genes from species that are stored in GenBank. Each unique sequence is ide ...
... This activity will allow you to use the tools used by geneticists and evolutionists located and maintained by the National Center for Biotechnology Information. These tools will allow you to compare the isolated and sequenced genes from species that are stored in GenBank. Each unique sequence is ide ...
A scale-free method for testing the proportionality of branch lengths
... change with time (but vary between genes) and if the two genes maintain their respective functions in the lineages under study, then the branch lengths of the phylogenetic tree for gene a (tree A) will most probably be different from the branch lengths of the phylogenetic tree for gene b (tree B); h ...
... change with time (but vary between genes) and if the two genes maintain their respective functions in the lineages under study, then the branch lengths of the phylogenetic tree for gene a (tree A) will most probably be different from the branch lengths of the phylogenetic tree for gene b (tree B); h ...
Algorithms and Data Analysis in Microarray Technology
... To recognize the biological information in data. To compare data from one array to another. In practice we do not understand the data – inevitably some biology will be removed too. ...
... To recognize the biological information in data. To compare data from one array to another. In practice we do not understand the data – inevitably some biology will be removed too. ...
DISEASES AND TREES - UC Berkeley College of Natural Resources
... Are my haplotypes sensitive enough? • To validate power of tool used, one needs to be able to differentiate among closely related individual • Generate progeny • Make sure each meiospore has different haplotype ...
... Are my haplotypes sensitive enough? • To validate power of tool used, one needs to be able to differentiate among closely related individual • Generate progeny • Make sure each meiospore has different haplotype ...
lecture notes
... • This allows each DNA site to choose its “favourite” tree in the network, independently of the others. • From a modelling perspective this is both a strength and a weakness (too liberal). • In any case: just solving the small parsimony problem on a network is, from a computational perspective, extr ...
... • This allows each DNA site to choose its “favourite” tree in the network, independently of the others. • From a modelling perspective this is both a strength and a weakness (too liberal). • In any case: just solving the small parsimony problem on a network is, from a computational perspective, extr ...
Fine scale mapping
... taking the value 1 if b has a parent in the gene tree and 0 otherwise. Allows for singleton leaf nodes, corresponding to sporadic case chromosomes, and disconnected sub-trees, corresponding to independent mutation events at the same disease locus. Assume number of branches of gene tree not removed i ...
... taking the value 1 if b has a parent in the gene tree and 0 otherwise. Allows for singleton leaf nodes, corresponding to sporadic case chromosomes, and disconnected sub-trees, corresponding to independent mutation events at the same disease locus. Assume number of branches of gene tree not removed i ...
PPTX - UT Computer Science
... Marker-based profiling can produce more accurate taxonomic profiles (distributions) than techniques that attempt to classify all fragments. ...
... Marker-based profiling can produce more accurate taxonomic profiles (distributions) than techniques that attempt to classify all fragments. ...
tutorialdm
... Each codon has 3 nucleotides, denote by fi (I = 1,2,3) Where s and n for a codon are given by s = ∑3i=1fi and n = (3-s) ...
... Each codon has 3 nucleotides, denote by fi (I = 1,2,3) Where s and n for a codon are given by s = ∑3i=1fi and n = (3-s) ...
Generating Workflow Graphs Using Typed Genetic Programming
... general enough to handle methods like k-means (where k affects the topology of the DAG) correctly, we had to use the polymorphic type “list of αs of size n” ([α]n ) with a natural number parameter n and an element type parameter α. The generating method systematically produces workflow DAGs from sim ...
... general enough to handle methods like k-means (where k affects the topology of the DAG) correctly, we had to use the polymorphic type “list of αs of size n” ([α]n ) with a natural number parameter n and an element type parameter α. The generating method systematically produces workflow DAGs from sim ...
No Slide Title - Faculty Virginia
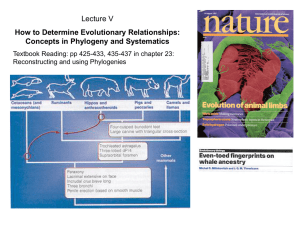
... •The tools of cladistics now represent the prevailing approach to determining relationships; the philosophy of strict monophyly wrt classification is still under debate -bears on definition - concept- of species ...
... •The tools of cladistics now represent the prevailing approach to determining relationships; the philosophy of strict monophyly wrt classification is still under debate -bears on definition - concept- of species ...
NSF I/UCRC Workshop Stony Brook University
... black dashed lines) - local constancy does not discourage long range interactions ...
... black dashed lines) - local constancy does not discourage long range interactions ...
What are Math and Computer Science doing in Biology?
... • A Tanglegram is a pair of phylogenetic trees drawn in the plane with no crossing edges, with the same labeled leaf set. The leaves of one tree are displayed on a line, and the leaves of the other tree are displayed on a parallel line. • One tree represents the evolution of a set of species, and th ...
... • A Tanglegram is a pair of phylogenetic trees drawn in the plane with no crossing edges, with the same labeled leaf set. The leaves of one tree are displayed on a line, and the leaves of the other tree are displayed on a parallel line. • One tree represents the evolution of a set of species, and th ...
Horizontal gene transfer and microbial evolution: Is the Tree-of
... find that tree that explains sequence data with minimum number of substitutions (tree includes hypothesis of sequence at each of the nodes) Maximum Likelihood analyses given a model for sequence evolution, find the tree that has the highest probability under this model. This approach can also be use ...
... find that tree that explains sequence data with minimum number of substitutions (tree includes hypothesis of sequence at each of the nodes) Maximum Likelihood analyses given a model for sequence evolution, find the tree that has the highest probability under this model. This approach can also be use ...
Statistical Methods for Genetic Association Mapping of Complex Traits with Related Individuals
... consider a general setting in which the complete data are dependent with marginal distributions following a generalized linear model. We form a vector Z whose elements are conditional expectations of the elements of the complete-data vector, given selected functions of the incomplete data. Assuming ...
... consider a general setting in which the complete data are dependent with marginal distributions following a generalized linear model. We form a vector Z whose elements are conditional expectations of the elements of the complete-data vector, given selected functions of the incomplete data. Assuming ...
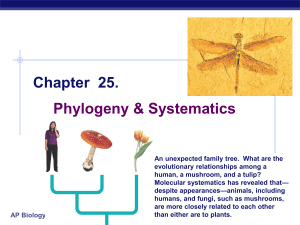
Ch_25 Phylogeny and Systematics
... genomes of different organisms, we find… humans & mice have 99% of their genes in ...
... genomes of different organisms, we find… humans & mice have 99% of their genes in ...
Matlab Bioinfo Toolbox QuickGuide
... MathWorks. It features a family of add-on application-specific solutions called toolboxes (i.e., comprehensive collections of functions) that extend the MATLAB environment to solve particular classes of problems. Bioinformatics Toolbox offers an integrated software environment for genome and proteom ...
... MathWorks. It features a family of add-on application-specific solutions called toolboxes (i.e., comprehensive collections of functions) that extend the MATLAB environment to solve particular classes of problems. Bioinformatics Toolbox offers an integrated software environment for genome and proteom ...
CHAPTER 24 Molecular Evolution
... d. Phenotypes can be misleading, because they do not always reflect genetic relatedness. i. Sometimes similarities result from convergent evolution, complicating the study of divergence among organisms (e.g., wings alone would put birds, bats and insects in the same evolutionary group). ii. Not all ...
... d. Phenotypes can be misleading, because they do not always reflect genetic relatedness. i. Sometimes similarities result from convergent evolution, complicating the study of divergence among organisms (e.g., wings alone would put birds, bats and insects in the same evolutionary group). ii. Not all ...
Analysis of Molecular Evolution in Mitochondrial tRNA Gene
... 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan. ...
... 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan. ...