Restriction fragment differential display of pediocin
... We have developed the so-called restriction fragment differential display PCR (RFDD-PCR) which is a variation of the cDNA amplified fragment length polymorphism DD technique originally described by Bachem et al. (1996). The technology is based on digesting cDNA with endonucleases followed by adaptor ...
... We have developed the so-called restriction fragment differential display PCR (RFDD-PCR) which is a variation of the cDNA amplified fragment length polymorphism DD technique originally described by Bachem et al. (1996). The technology is based on digesting cDNA with endonucleases followed by adaptor ...
CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE
... • If the genes were completely linked, expected results from the testcross would be a 1:1 phenotypic ratio of parental types only. • Morgan's testcross did not produce results consistent with unlinkage or total linkage. The high proportion of parental phenotypes suggested linkage between the two gen ...
... • If the genes were completely linked, expected results from the testcross would be a 1:1 phenotypic ratio of parental types only. • Morgan's testcross did not produce results consistent with unlinkage or total linkage. The high proportion of parental phenotypes suggested linkage between the two gen ...
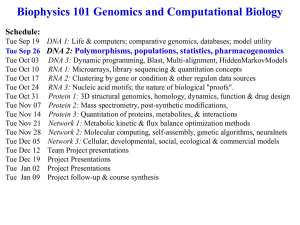
Biophysics 101 Genomics and Computational Biology
... Isolation and properties of Escherichia coli ATPase mutants with altered divalent metal specificity for ATP hydrolysis. Isolation of altered specificity mutants of the single-chain 434 repressor that recognize asymmetric DNA sequences containing TTAA Mechanisms of spontaneous mutagenesis: clues from ...
... Isolation and properties of Escherichia coli ATPase mutants with altered divalent metal specificity for ATP hydrolysis. Isolation of altered specificity mutants of the single-chain 434 repressor that recognize asymmetric DNA sequences containing TTAA Mechanisms of spontaneous mutagenesis: clues from ...
Two groups of human herpesvirus 6 identified by sequence
... As HHV-6 has a tropism similar to that of human immunodeficiency virus (HIV) in infections of CD4 ÷ T lymphocytes and monocytes/macrophages (Yamanishi et al., 1988; Okuno et al., 1989; Levy et al., 1990a; Kondo et al., 1991; Wrzos et al., 1990), it has been suggested that HHV-6 may reactivate and pa ...
... As HHV-6 has a tropism similar to that of human immunodeficiency virus (HIV) in infections of CD4 ÷ T lymphocytes and monocytes/macrophages (Yamanishi et al., 1988; Okuno et al., 1989; Levy et al., 1990a; Kondo et al., 1991; Wrzos et al., 1990), it has been suggested that HHV-6 may reactivate and pa ...
Parallel Genetic and Phenotypic Evolution of DNA Superhelicity in
... found in topA and fis and, by using isogenic strains, it was demonstrated that these two mutations were responsible for the observed changes in DNA superhelicity in that population and, moreover, both were beneficial under the conditions of the evolution experiment. However, that previous study did ...
... found in topA and fis and, by using isogenic strains, it was demonstrated that these two mutations were responsible for the observed changes in DNA superhelicity in that population and, moreover, both were beneficial under the conditions of the evolution experiment. However, that previous study did ...
In vitro conjugal transfer of tetracycline resistance from Lactobacillus
... 512). Based on our conjugation experiments with only one S. aureus strain, we cannot exclude this species from the host range. In a few transconjugants (TC 500-3, TC 507-1, TC 5074 and TC 515-1), additional plasmids, other than the plasmid encoding the tetracycline resistance, seemed to have co-tran ...
... 512). Based on our conjugation experiments with only one S. aureus strain, we cannot exclude this species from the host range. In a few transconjugants (TC 500-3, TC 507-1, TC 5074 and TC 515-1), additional plasmids, other than the plasmid encoding the tetracycline resistance, seemed to have co-tran ...
Identification and Analysis of Arabidopsis Expressed Sequence
... a family of ncRNAs with members in several plant species (Taylor and Green, 1995; Teramoto et al., 1996; van Hoof et al., 1997). GUT15 (gene with unstable transcript 15) is another characterized member of this family. This transcript was first identified as one of the most unstable transcripts in to ...
... a family of ncRNAs with members in several plant species (Taylor and Green, 1995; Teramoto et al., 1996; van Hoof et al., 1997). GUT15 (gene with unstable transcript 15) is another characterized member of this family. This transcript was first identified as one of the most unstable transcripts in to ...
Answer Appendix B - McGraw Hill Higher Education
... sequence of DNA within that polymer; the sequence of bases creates a gene and distinguishes it from other genes. Genes are located in chromosomes, which are found within living cells. C4. At the molecular level, a gene (a sequence of DNA) is first transcribed into RNA. The genetic code within the RN ...
... sequence of DNA within that polymer; the sequence of bases creates a gene and distinguishes it from other genes. Genes are located in chromosomes, which are found within living cells. C4. At the molecular level, a gene (a sequence of DNA) is first transcribed into RNA. The genetic code within the RN ...
The Rat Gene Map
... segments found when either species is compared with human. It should be kept in mind that these calculations are very approximate due to the scarcity of data, but they might still provide hints of genomic relationships among the 3 species. Thus, what appears to be a considerable amount of genome rea ...
... segments found when either species is compared with human. It should be kept in mind that these calculations are very approximate due to the scarcity of data, but they might still provide hints of genomic relationships among the 3 species. Thus, what appears to be a considerable amount of genome rea ...
What`s new - JSI medical systems
... The entry in Suffix can have up to six characters. The suffix is added to the Name of the ROI, e.g: Suffix ng (for NimbleGene), ROI name is BRCA1E01-ng. Therefore all ROIs added on tab Enrichment have a suffix in the ROI name listed in column Name. There is the setting auto cut available: If you use ...
... The entry in Suffix can have up to six characters. The suffix is added to the Name of the ROI, e.g: Suffix ng (for NimbleGene), ROI name is BRCA1E01-ng. Therefore all ROIs added on tab Enrichment have a suffix in the ROI name listed in column Name. There is the setting auto cut available: If you use ...
Transposable elements activity reveals punctuated
... new parameter called Density of Insertion (DI), which is the ratio between the number of TE insertions in a genome and its size. We calculated the DI at both divergence thresholds (1%DI and 5%DI). As for mammalian speciation patterns, we also calculated the Rate of Speciation (RS) as the ratio betwe ...
... new parameter called Density of Insertion (DI), which is the ratio between the number of TE insertions in a genome and its size. We calculated the DI at both divergence thresholds (1%DI and 5%DI). As for mammalian speciation patterns, we also calculated the Rate of Speciation (RS) as the ratio betwe ...
Construction of in vivo infectious clones of PRSV P-YK and W-CI
... However, WMV-1 and PRSV isolates are serologically indistinguishable when tested against antibodies prepared to viral particles (Gonsalves & Ishii, 1980), coat protein (CP) (Yeh et al., 1984), cylindrical inclusion proteins (CIPs) (Yeh & Gonsalves, 1984b), and ...
... However, WMV-1 and PRSV isolates are serologically indistinguishable when tested against antibodies prepared to viral particles (Gonsalves & Ishii, 1980), coat protein (CP) (Yeh et al., 1984), cylindrical inclusion proteins (CIPs) (Yeh & Gonsalves, 1984b), and ...
Rearrangements in the Human T-Cell-Receptor Â
... ATL3 is defined on the basis of the clinical picture, distinct morphology of the abnormal lymphocytes, and T-cell markers. Particularly close association of HTLV-I infection with leukemogenesis of ATL has often been reported by a number of serological and epidemiológica! studies (1,2). However, lac ...
... ATL3 is defined on the basis of the clinical picture, distinct morphology of the abnormal lymphocytes, and T-cell markers. Particularly close association of HTLV-I infection with leukemogenesis of ATL has often been reported by a number of serological and epidemiológica! studies (1,2). However, lac ...
Epidemiology and control of fungally transmitted cereal viruses
... barley mild mosaic virus (BaMMV) and barley yellow mosaic virus (BaYMV) and their fungus vector, Polymyxa graminis. This includes studies of vector biology, variability and control; the viruses and their variability; methods for diagnosis of virus in soil and the nature of cultivar resistance. Studi ...
... barley mild mosaic virus (BaMMV) and barley yellow mosaic virus (BaYMV) and their fungus vector, Polymyxa graminis. This includes studies of vector biology, variability and control; the viruses and their variability; methods for diagnosis of virus in soil and the nature of cultivar resistance. Studi ...
DNA sequencing revealed a definitive
... Figure 1. Restriction maps of the regions of XHB3 and XHB4 containing the sea urchin homeo boxes. Horizontal arrows on top show the sequencing strategy. All sequencing was done by the dideoxy chain termination method (19) after subcloning the restriction fragments into M13. Dale deletion subclones f ...
... Figure 1. Restriction maps of the regions of XHB3 and XHB4 containing the sea urchin homeo boxes. Horizontal arrows on top show the sequencing strategy. All sequencing was done by the dideoxy chain termination method (19) after subcloning the restriction fragments into M13. Dale deletion subclones f ...
evolution and mechanism of translation in chloroplasts
... Chloroplasts are plant organelles that contain the entire machinery for the process of photosynthesis. In addition, chloroplasts possess their own genome, multiple copies of circular double-stranded DNA molecules, typically 150 kb in size, with over 100 different genes. According to the endosymbioti ...
... Chloroplasts are plant organelles that contain the entire machinery for the process of photosynthesis. In addition, chloroplasts possess their own genome, multiple copies of circular double-stranded DNA molecules, typically 150 kb in size, with over 100 different genes. According to the endosymbioti ...
and Mountain Lions (Puma concolor) in North America
... 500 to 700 bp (see Table SA2 and Fig. SA1 in the supplemental material). To minimize sequencing errors, we sought to achieve at least 2⫻ sequencing coverage at all positions in each viral genome. However, small regions in some viral genomes with high-quality chromatograms had single coverage. All ch ...
... 500 to 700 bp (see Table SA2 and Fig. SA1 in the supplemental material). To minimize sequencing errors, we sought to achieve at least 2⫻ sequencing coverage at all positions in each viral genome. However, small regions in some viral genomes with high-quality chromatograms had single coverage. All ch ...
Bioinformatics with basic local alignment search tool (BLAST) and
... such as Protein Data Bank. Secondly, bioinformatics seeks to develop tools for analysis of data, and thirdly, to use these tools to analyze and interpret data results, which is the focus of the current review. In recent times, application of computational approaches to biological data has become a v ...
... such as Protein Data Bank. Secondly, bioinformatics seeks to develop tools for analysis of data, and thirdly, to use these tools to analyze and interpret data results, which is the focus of the current review. In recent times, application of computational approaches to biological data has become a v ...
Staphylococcus aureus CC395 harbours a novel
... Conclusions: The size and complexity of the SCCmec element support the idea that CC395 is highly prone to DNA uptake from CoNS. Thus CC395 may serve as an entry point for SCCmec and SCC structures into S. aureus. ...
... Conclusions: The size and complexity of the SCCmec element support the idea that CC395 is highly prone to DNA uptake from CoNS. Thus CC395 may serve as an entry point for SCCmec and SCC structures into S. aureus. ...
Complete
... It has been proposed that an array of spatially asymmetric obstacles could operate as Brownian ratchets, structures that permit Brownian motion in only one direction [1–7]. When particles flow through such an array driven by an electric field (Fig. 2.1A), particles diffusing to the left (path 1; Fig ...
... It has been proposed that an array of spatially asymmetric obstacles could operate as Brownian ratchets, structures that permit Brownian motion in only one direction [1–7]. When particles flow through such an array driven by an electric field (Fig. 2.1A), particles diffusing to the left (path 1; Fig ...
Genomic library

A genomic library is a collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.There are several kinds of vectors available with various insert capacities. Generally, libraries made from organisms with larger genomes require vectors featuring larger inserts, thereby fewer vector molecules are needed to make the library. Researchers can choose a vector also considering the ideal insert size to find a desired number of clones necessary for full genome coverage.Genomic libraries are commonly used for sequencing applications. They have played an important role in the whole genome sequencing of several organisms, including the human genome and several model organisms.