BLAST seminar
... • Scoring scheme for quality of an alignment • Contains scores for every possible amino acid substitution in a sequence alignment • For protein/protein comparisons we need a 20 x 20 matrix with scores for pairs of residues. Every cell in the matrix contains at position X, Y a score for the substitut ...
... • Scoring scheme for quality of an alignment • Contains scores for every possible amino acid substitution in a sequence alignment • For protein/protein comparisons we need a 20 x 20 matrix with scores for pairs of residues. Every cell in the matrix contains at position X, Y a score for the substitut ...
n 1 , n 2 , n 3 - Carnegie Mellon School of Computer Science
... The first model is designed for analyses of conserved linkage of genes in three regions from three distinct genomes. The second model is for detection of segments duplicated by a whole genome duplication (WGD), via comparison with the genome of a related, pre-duplication species. We again use a Venn ...
... The first model is designed for analyses of conserved linkage of genes in three regions from three distinct genomes. The second model is for detection of segments duplicated by a whole genome duplication (WGD), via comparison with the genome of a related, pre-duplication species. We again use a Venn ...
Evidence That Plant-Like Genes in Chlamydia Species Reflect an
... However, such analysis does not group these lineages with high confidence. This is most likely due to a significant divergence time between these lineages, which severely limits the phylogenetic information (informative sites) available, and also reduces the number of gene sequences that can be anal ...
... However, such analysis does not group these lineages with high confidence. This is most likely due to a significant divergence time between these lineages, which severely limits the phylogenetic information (informative sites) available, and also reduces the number of gene sequences that can be anal ...
Evidence That Plant-Like Genes in Chlamydia
... However, such analysis does not group these lineages with high confidence. This is most likely due to a significant divergence time between these lineages, which severely limits the phylogenetic information (informative sites) available, and also reduces the number of gene sequences that can be anal ...
... However, such analysis does not group these lineages with high confidence. This is most likely due to a significant divergence time between these lineages, which severely limits the phylogenetic information (informative sites) available, and also reduces the number of gene sequences that can be anal ...
Real time PCR or Quantitative PCR
... ○ Bernard and Wittwer (2002) used real-time PCR for detection of multiple breast cancer molecular markers ● Genetic variation analysis ...
... ○ Bernard and Wittwer (2002) used real-time PCR for detection of multiple breast cancer molecular markers ● Genetic variation analysis ...
Snork Activity
... peptide bonds and form a polypeptide. The process in which the original DNA information (carried by the mRNA) is transferred into a protein is called translation. In this investigation you will simulate the process of transcription and translation to create the proteins that will develop the charact ...
... peptide bonds and form a polypeptide. The process in which the original DNA information (carried by the mRNA) is transferred into a protein is called translation. In this investigation you will simulate the process of transcription and translation to create the proteins that will develop the charact ...
Deciphering the molecular phylogenetics of the Asian honey bee
... The BLAST analysis of 513 bp of the insect A. cerana showed significant homology with A. cerana from Indonesia. The phylogenetic NJ tree was carried out using MEGA6 software. The NJ tree was constructed based on the multiple aligned sequence data for different Apis species. The tree separates the ge ...
... The BLAST analysis of 513 bp of the insect A. cerana showed significant homology with A. cerana from Indonesia. The phylogenetic NJ tree was carried out using MEGA6 software. The NJ tree was constructed based on the multiple aligned sequence data for different Apis species. The tree separates the ge ...
The Functional Organization of the Vestigial Locus in Drosophila
... (described in Lindsley and Grell 1968; Williams and Bell 1988). Bownes and Roberts (1981b) have proposed that the cell death observed in vg mutants may be the consequence of abnormal positional information in vg wing imaginal discs. Thus, a molecular analysis of vg is important since it may help to ...
... (described in Lindsley and Grell 1968; Williams and Bell 1988). Bownes and Roberts (1981b) have proposed that the cell death observed in vg mutants may be the consequence of abnormal positional information in vg wing imaginal discs. Thus, a molecular analysis of vg is important since it may help to ...
Fast, simultaneous, and sensitive detection of staphylococci
... amounts of DNA from Gram positive bacteria. DNA from staphylococci is notoriously difficult to isolate because of the membrane structure of these organisms.4 We have shown that lysis in an SDS/β mercaptoethanol buffer and exposure to microwaves is as effective as a longer, more elaborate, and more e ...
... amounts of DNA from Gram positive bacteria. DNA from staphylococci is notoriously difficult to isolate because of the membrane structure of these organisms.4 We have shown that lysis in an SDS/β mercaptoethanol buffer and exposure to microwaves is as effective as a longer, more elaborate, and more e ...
Introduction to Bioinformatics
... The purpose of this exercise is to introduce to you and familiarize you with the National Center for Biotechnology Information (NCBI) web sites. These web sites contain nucleotide and protein databases containing a vast amount of sequencing data that is increasing every single day. As more and more ...
... The purpose of this exercise is to introduce to you and familiarize you with the National Center for Biotechnology Information (NCBI) web sites. These web sites contain nucleotide and protein databases containing a vast amount of sequencing data that is increasing every single day. As more and more ...
METADOMAIN: A PROFILE HMM-BASED PROTEIN DOMAIN
... As both the reference genome and its domain annotations are available, we can quantify the sensitivity and false positive (FP) rate of MetaDomain. The second one is metagenome data sequenced from soil. We applied MetaDomain to identify domains encoded in the underlying data. In addition, we compared ...
... As both the reference genome and its domain annotations are available, we can quantify the sensitivity and false positive (FP) rate of MetaDomain. The second one is metagenome data sequenced from soil. We applied MetaDomain to identify domains encoded in the underlying data. In addition, we compared ...
Bio212-01-Alu Lab Part1
... Over time, many short inserted sequences have accumulated in our DNA. For example, most of us carry approximately 500,000 copies of a 300 bp sequence known as the Alu sequence in our DNA. The origin and function of these sequences are still unknown. Despite this, these repeated Alu sequences have pr ...
... Over time, many short inserted sequences have accumulated in our DNA. For example, most of us carry approximately 500,000 copies of a 300 bp sequence known as the Alu sequence in our DNA. The origin and function of these sequences are still unknown. Despite this, these repeated Alu sequences have pr ...
Gene Section IGK (Immunoglobulin Kappa) Atlas of Genetics and Cytogenetics
... IGKJ segments, and a unique IGKC gene. The 76 IGKV genes are organized in two clusters separated by 800 kb. The IGKV distal cluster (the most 5' from IGKC and in the most centromeric position) spans 400 kb and comprises 36 genes. The IGKV proximal cluster (in 3' of the locus, closer to IGKC, and in ...
... IGKJ segments, and a unique IGKC gene. The 76 IGKV genes are organized in two clusters separated by 800 kb. The IGKV distal cluster (the most 5' from IGKC and in the most centromeric position) spans 400 kb and comprises 36 genes. The IGKV proximal cluster (in 3' of the locus, closer to IGKC, and in ...
ANNEX 1
... The Departments/Laboratories shown in annexes were assessed according to the Accreditation Criteria for Medical Laboratories, as defined in the Standard ...
... The Departments/Laboratories shown in annexes were assessed according to the Accreditation Criteria for Medical Laboratories, as defined in the Standard ...
genomic library
... • Restriction enzymes cut DNA into specific fragments • Restriction enzymes recognize specific base sequences in double-stranded DNA and cleave both strands of the duplex at specific places • Characteristics of restriction enzymes: 1. Cut DNA sequence-specifically 2. Bacterial enzymes; hundreds are ...
... • Restriction enzymes cut DNA into specific fragments • Restriction enzymes recognize specific base sequences in double-stranded DNA and cleave both strands of the duplex at specific places • Characteristics of restriction enzymes: 1. Cut DNA sequence-specifically 2. Bacterial enzymes; hundreds are ...
Laboratory B - Filogeografía
... Lab 2, page 5 of 6 Now select File > Export . . . twice, choosing different formats each time, such as FASTA and “OldFashioned NEXUS”, just to be safe, and perhaps adding “revcomp” to the end of the name. Save data file as well, under a new name. Open one of these new files in MEGA, and follow the i ...
... Lab 2, page 5 of 6 Now select File > Export . . . twice, choosing different formats each time, such as FASTA and “OldFashioned NEXUS”, just to be safe, and perhaps adding “revcomp” to the end of the name. Save data file as well, under a new name. Open one of these new files in MEGA, and follow the i ...
Comparative Genomics Reveals Adaptive Protein Evolution and a
... mordax, Oncorhynchus mykiss, Ictalurus punctatus, and Esox lucius) were retrieved from BLASTX outputs to be used as outgroups. We inferred the most parsimonious ancestral state at each polymorphic site in eels by aligning orthologous genes based on their translated sequences using CLUSTALW (Thompson ...
... mordax, Oncorhynchus mykiss, Ictalurus punctatus, and Esox lucius) were retrieved from BLASTX outputs to be used as outgroups. We inferred the most parsimonious ancestral state at each polymorphic site in eels by aligning orthologous genes based on their translated sequences using CLUSTALW (Thompson ...
Gene finding: putting the parts together
... Any isolated signal of a gene is hard to predict. Current methods for promoter prediction, for instance, will have either a very low specificity or a very bad sensitivity, such that they will either predict a huge number of false positives (fake promoters) or a very small number of true promoters. T ...
... Any isolated signal of a gene is hard to predict. Current methods for promoter prediction, for instance, will have either a very low specificity or a very bad sensitivity, such that they will either predict a huge number of false positives (fake promoters) or a very small number of true promoters. T ...
Comprehensive Cardiomyopathy Panel
... ordered as a cost-effective reflex following targeted disease testing (DCM, HCM, LVNC, or ...
... ordered as a cost-effective reflex following targeted disease testing (DCM, HCM, LVNC, or ...
Enter Topic Title in each section above
... is described as being pathogenic? A. Disease-causing Q. Describe how some bacteria respond in order to survive when environmental conditions become unfavourable. A. Produce (endo)spores ...
... is described as being pathogenic? A. Disease-causing Q. Describe how some bacteria respond in order to survive when environmental conditions become unfavourable. A. Produce (endo)spores ...
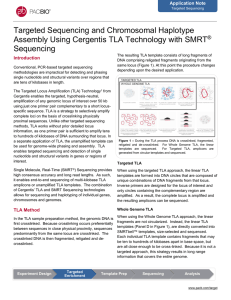
Application Note: Targeted sequencing and chromosomal haplotype
... with ~4 segments per read. The schematic in Figure 3 illustrates the general structure of the reads and how they mapped to the reference. Mapped reads generated with the Targeted TLA to BRCA1 fully cover the BRCA1 region (Figure 4), with heterozygous SNPs clearly visible from the TLA data (Figure 5) ...
... with ~4 segments per read. The schematic in Figure 3 illustrates the general structure of the reads and how they mapped to the reference. Mapped reads generated with the Targeted TLA to BRCA1 fully cover the BRCA1 region (Figure 4), with heterozygous SNPs clearly visible from the TLA data (Figure 5) ...
Analysing thousands of bacterial genomes: gene annotation
... Interpretation: this shows that with increase in –depth level, you select more number of species for phylogenetic profiles which sometime leads to over-representation of particular taxon, hence it may bias the result. Increase in –depth level also increases the program execution time. ...
... Interpretation: this shows that with increase in –depth level, you select more number of species for phylogenetic profiles which sometime leads to over-representation of particular taxon, hence it may bias the result. Increase in –depth level also increases the program execution time. ...
Concept_Paper
... Alveolate clade to have a genome sequence. Complete genome information should facilitate investigations of the biology, not just of other ciliate model organisms, but also of a variety of organisms of medical and agricultural importance. Experimental in vivo functional genomics: one of the most va ...
... Alveolate clade to have a genome sequence. Complete genome information should facilitate investigations of the biology, not just of other ciliate model organisms, but also of a variety of organisms of medical and agricultural importance. Experimental in vivo functional genomics: one of the most va ...
Specific oligonucleotide primers for detection of endoglucanase
... endoglucanase gene of B. subtilis, G. stearothermophilus and P. campinasensis. Two PCR primers, EN1F and EN1R, were chosen that were predicted to specifically amplify a 1,311 bpDNA fragment of the B. Subtilis, G. stearothermophilus and P. campinasensis. The Genbank database (NCBI) search for complim ...
... endoglucanase gene of B. subtilis, G. stearothermophilus and P. campinasensis. Two PCR primers, EN1F and EN1R, were chosen that were predicted to specifically amplify a 1,311 bpDNA fragment of the B. Subtilis, G. stearothermophilus and P. campinasensis. The Genbank database (NCBI) search for complim ...
Study of Hypertension in Spontaneous Hypertensive Rats by
... Results: There were no important differences between all the above rats in the 6,268 nucleotides of the alpha2B receptor. Conclusion: Our data suggest that, although the alpha2B adrenergic receptor has a crucial role in the development of hypertension, the genetic susceptibility or resistance of var ...
... Results: There were no important differences between all the above rats in the 6,268 nucleotides of the alpha2B receptor. Conclusion: Our data suggest that, although the alpha2B adrenergic receptor has a crucial role in the development of hypertension, the genetic susceptibility or resistance of var ...
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Recent studies use either ""shotgun"" or PCR directed sequencing to get largely unbiased samples of all genes from all the members of the sampled communities. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale and detail than before.