Chapter 2 - SCHOOLinSITES
... – Substrate binds to enzyme at active site – Enzymes act on substrates to reduce energy needed to make product – Substrate is changed – Enzyme separates from products and can form an association with another substrate – Enzyme, as a catalyst is not used up in the reaction – Increases reaction rate ...
... – Substrate binds to enzyme at active site – Enzymes act on substrates to reduce energy needed to make product – Substrate is changed – Enzyme separates from products and can form an association with another substrate – Enzyme, as a catalyst is not used up in the reaction – Increases reaction rate ...
This exam has 9 pages, including this one.
... i) A protein that is 20 amino acid residues in length folds into a stable structure. Assume that the protein forms all but one hydrogen bond when it folds and that the unsatisfied H-bond is not accessible to water. Calculate the enthalpy of unfolding. State whatever assumptions you make regarding th ...
... i) A protein that is 20 amino acid residues in length folds into a stable structure. Assume that the protein forms all but one hydrogen bond when it folds and that the unsatisfied H-bond is not accessible to water. Calculate the enthalpy of unfolding. State whatever assumptions you make regarding th ...
Corn Bt11 x DA59122 x MIR604 x TC1507 x GA21
... Determination of the Safety of Syngenta’s Combined trait product corn: Bt11 x DAS59122 xMIR604 x TC1507 x GA21 forDirect use as Food, Feed, or Processing Food and Feed Safety The product dossier on Syngenta’scombined trait product corn: Bt11 x DAS59122 x MIR604 x TC1507 x GA21 was reviewed for safet ...
... Determination of the Safety of Syngenta’s Combined trait product corn: Bt11 x DAS59122 xMIR604 x TC1507 x GA21 forDirect use as Food, Feed, or Processing Food and Feed Safety The product dossier on Syngenta’scombined trait product corn: Bt11 x DAS59122 x MIR604 x TC1507 x GA21 was reviewed for safet ...
This exam has 9 pages, including this one.
... B6: (8 pts) The core of a protein contains a Valine residue in the wild-type enzyme. This is replaced by Leucine in one mutant and Threonine in another. The thermodynamic parameter associated with unfolding of each of these proteins is provided below the amino acid side-chain. The direction of the r ...
... B6: (8 pts) The core of a protein contains a Valine residue in the wild-type enzyme. This is replaced by Leucine in one mutant and Threonine in another. The thermodynamic parameter associated with unfolding of each of these proteins is provided below the amino acid side-chain. The direction of the r ...
Ruboyianes - University of Arizona
... protein is ejected from the capsid along with the penetrating DNA (14). Initially, protein H and the incoming DNA are associated with the outer membrane (13) at sites of cell wall adhesion (2, 3). From there, the single-stranded DNA (ssDNA) is delivered to the cytoplasmic membrane, the site of DNA s ...
... protein is ejected from the capsid along with the penetrating DNA (14). Initially, protein H and the incoming DNA are associated with the outer membrane (13) at sites of cell wall adhesion (2, 3). From there, the single-stranded DNA (ssDNA) is delivered to the cytoplasmic membrane, the site of DNA s ...
Protein-only inheritance in yeast: something to get
... experiments analysing the effects of point mutations in Hsp104. Hsp104 contains two Walker-type nucleotide-binding sites that are crucial for its function in thermotolerance in yeast36. Mutation of both of these sites cures [PSI1], and fluorescence from Sup35p–GFP fusion proteins is diffuse in these ...
... experiments analysing the effects of point mutations in Hsp104. Hsp104 contains two Walker-type nucleotide-binding sites that are crucial for its function in thermotolerance in yeast36. Mutation of both of these sites cures [PSI1], and fluorescence from Sup35p–GFP fusion proteins is diffuse in these ...
Word file - UC Davis
... Based on the arrows representing the strands, Nter is at 2, and 1 is Cter. 7) The so-called Rosetta stone for predicting protein-protein interactions is: A) Gene fusion B) Gene co-expression C) Presence of the name of the two proteins concerned in the same scientific paper D) A very old stone recent ...
... Based on the arrows representing the strands, Nter is at 2, and 1 is Cter. 7) The so-called Rosetta stone for predicting protein-protein interactions is: A) Gene fusion B) Gene co-expression C) Presence of the name of the two proteins concerned in the same scientific paper D) A very old stone recent ...
Spnr, a Murine RNA-binding Protein That Is Localized to
... mRNA stability, transport, localization, and translation (Jackson, 1993). Although antisense RNAs and proteins that bind to the 3' UTRs of some mRNAs have been identified (Wightman et al., 1993; Lee et al., 1993; Wharton and Struhl, 1991; Harford et al., 1990), the molecular mechanisms by which 3' U ...
... mRNA stability, transport, localization, and translation (Jackson, 1993). Although antisense RNAs and proteins that bind to the 3' UTRs of some mRNAs have been identified (Wightman et al., 1993; Lee et al., 1993; Wharton and Struhl, 1991; Harford et al., 1990), the molecular mechanisms by which 3' U ...
Quant-iT™ Assay Kits for microplate
... of RNA. The x-axis gives the mass of nucleic acid when DNA or RNA is assayed alone; in the 1:1 mixture, the total mass of nucleic acid is double the amount shown. The inset shows the sensitivity of the assay for DNA. B The Quant-iT™ RNA Assay Kit has a linear detection range of 5–100 ng and is selec ...
... of RNA. The x-axis gives the mass of nucleic acid when DNA or RNA is assayed alone; in the 1:1 mixture, the total mass of nucleic acid is double the amount shown. The inset shows the sensitivity of the assay for DNA. B The Quant-iT™ RNA Assay Kit has a linear detection range of 5–100 ng and is selec ...
Reece9e_Lecture_C05
... protein under normal cellular conditions. A protein’s specific structure determines its function. When a cell synthesizes a polypeptide, the chain generally folds spontaneously to assume the functional structure for that protein. The folding is reinforced by a variety of bonds between parts of ...
... protein under normal cellular conditions. A protein’s specific structure determines its function. When a cell synthesizes a polypeptide, the chain generally folds spontaneously to assume the functional structure for that protein. The folding is reinforced by a variety of bonds between parts of ...
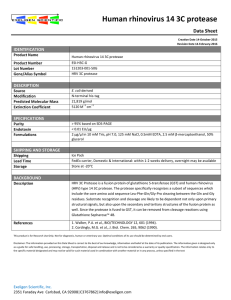
HRV_3C_protease_PDS_V1.0
... include the core amino acid sequence Leu-‐Phe-‐Gln/Gly-‐Pro cleaving between the Gln and Gly residues. Substrate recognition and cleavage are likely to be dependent not only upon primary structural signals, b ...
... include the core amino acid sequence Leu-‐Phe-‐Gln/Gly-‐Pro cleaving between the Gln and Gly residues. Substrate recognition and cleavage are likely to be dependent not only upon primary structural signals, b ...
... This exam consists of 5 pages and 15 questions. Total points are 100. Allot 1 min/2 points. On questions with choices, all of your answers will be graded and the best scoring answer will be used. Use the space provided, or the back of the previous page. The full name and three letter abbreviation of ...
Proteins – Essential Biomolecules
... Faulty protein production has many potential causes. The gene base for each specific protein is called a gene. of such projects and they all involve skilled science workers. Lipids act as thecould buildingbe blocks for cell membranes by organising into a double sequence wrong. The process of gene exp ...
... Faulty protein production has many potential causes. The gene base for each specific protein is called a gene. of such projects and they all involve skilled science workers. Lipids act as thecould buildingbe blocks for cell membranes by organising into a double sequence wrong. The process of gene exp ...
462a Reading and Homework Assignment 3
... The histidine that is hydrogen bonded to the inhibitor in the TIM complex is residue number 95. (b) What is the three-letter group name (that is, the “residue” name) for the inhibitor, 2phosphoglycolate, bound to TIM in this structure? (Hint: click on the inhibitor). PGA (7) Examine the two Ramachan ...
... The histidine that is hydrogen bonded to the inhibitor in the TIM complex is residue number 95. (b) What is the three-letter group name (that is, the “residue” name) for the inhibitor, 2phosphoglycolate, bound to TIM in this structure? (Hint: click on the inhibitor). PGA (7) Examine the two Ramachan ...
Online Counseling Resource YCMOU ELearning Drive…
... electrically charged. These interact with each other and their surroundings in the cell to produce a well-defined, three dimensional shape, the folded protein., known as the native state. The resulting three-dimensional structure is determined by the sequence of the amino acids. © 2008, YCMOU. A ...
... electrically charged. These interact with each other and their surroundings in the cell to produce a well-defined, three dimensional shape, the folded protein., known as the native state. The resulting three-dimensional structure is determined by the sequence of the amino acids. © 2008, YCMOU. A ...
aminoacyl-tRNA synthetases
... ribosome translocation as prokaryotes • there is no E site on eukaryotic ribosomes, only A and ...
... ribosome translocation as prokaryotes • there is no E site on eukaryotic ribosomes, only A and ...
Untitled
... Hershey and Chase grew one batch of E. coli in a medium containing 32P and infected the bacteria with T2 phage so that all the new phages would have DNA labeled with 32P (Figure 10.5). They grew a second batch of E. coli in a medium containing 35S and infected these bacteria with T2 phage so that ...
... Hershey and Chase grew one batch of E. coli in a medium containing 32P and infected the bacteria with T2 phage so that all the new phages would have DNA labeled with 32P (Figure 10.5). They grew a second batch of E. coli in a medium containing 35S and infected these bacteria with T2 phage so that ...
The chemical constituents of cells
... Being amphoteric amino acids can act as buffer solutions. A buffer solution is one which resists the tendency to change its pH even when small amounts of acids or alkali are added to it. Such a property is esstenial in biological systems where any sudden change in pH could adversely affect the funct ...
... Being amphoteric amino acids can act as buffer solutions. A buffer solution is one which resists the tendency to change its pH even when small amounts of acids or alkali are added to it. Such a property is esstenial in biological systems where any sudden change in pH could adversely affect the funct ...
Amino acid sequence restriction in relation to proteolysis
... The d a t a in T a b l e s 1-5 suggest a role of proline residues in regulation of proteolysis. Proline is over-represented at potentially s e n s i t i v e s i t e s in regions not s t a b i l i z e d by secondary structure. C o n s e q u e n t l y , dibasic structures not stabilized by either seco ...
... The d a t a in T a b l e s 1-5 suggest a role of proline residues in regulation of proteolysis. Proline is over-represented at potentially s e n s i t i v e s i t e s in regions not s t a b i l i z e d by secondary structure. C o n s e q u e n t l y , dibasic structures not stabilized by either seco ...
Two-hybrid screening
Two-hybrid screening (also known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively.The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the binding domain (BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay.