Method S1.
... mM Tris-HCl, 100 mM NaCl (pH 7.4; final volume 1 ml). Absorbance at 340 nm ( = 6220 M-1 cm-1) was recorded for 3 min and subtracted from the absorbance of a control reaction carried out omitting NAD+. In order to validate the method, control reactions were performed replacing the sample with L-glut ...
... mM Tris-HCl, 100 mM NaCl (pH 7.4; final volume 1 ml). Absorbance at 340 nm ( = 6220 M-1 cm-1) was recorded for 3 min and subtracted from the absorbance of a control reaction carried out omitting NAD+. In order to validate the method, control reactions were performed replacing the sample with L-glut ...
Macromolecules: Proteins
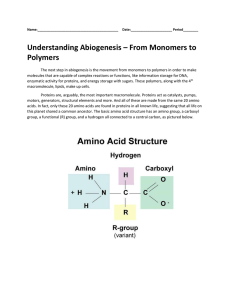
... Color code the amino acid on this worksheet (carbon-black, hydrogen-yellow, nitrogen-blue, and oxygen-red). Basic Structure of Amino acid H ...
... Color code the amino acid on this worksheet (carbon-black, hydrogen-yellow, nitrogen-blue, and oxygen-red). Basic Structure of Amino acid H ...
XL-I
... Hb demonstrates higher oxygen carrying capacity compared to myoglobin II. There is covalent bonding between the four subunits of Hb III. During deoxygenation the loss of the first oxygen molecule from oxygenated Hb promotes the dissociation of oxygen from the other subunits (A) II ...
... Hb demonstrates higher oxygen carrying capacity compared to myoglobin II. There is covalent bonding between the four subunits of Hb III. During deoxygenation the loss of the first oxygen molecule from oxygenated Hb promotes the dissociation of oxygen from the other subunits (A) II ...
McPherson, Selwyn-Lloyd: Investigations Into a Genetic Algorithm for Protein Sequences
... more helpful, but with so little information to mutate, the solution space becomes too coarse and thus more random, hindering the smooth traversal of the solution space. Fortunately, by bringing the fundamental concepts of the GA back to their natural environment, these problems are alleviated! Chro ...
... more helpful, but with so little information to mutate, the solution space becomes too coarse and thus more random, hindering the smooth traversal of the solution space. Fortunately, by bringing the fundamental concepts of the GA back to their natural environment, these problems are alleviated! Chro ...
ppt - University of Illinois Urbana
... – P(X|Y)=(count(X;Y)+pseudocount(X;Y) )/(count(Y)+totalpseudocount) – E.g., p(v|M1)=(“count of v in M1 column”+1)/ (“all amino acids in M1 column”+20) (Laplace smoothing) ...
... – P(X|Y)=(count(X;Y)+pseudocount(X;Y) )/(count(Y)+totalpseudocount) – E.g., p(v|M1)=(“count of v in M1 column”+1)/ (“all amino acids in M1 column”+20) (Laplace smoothing) ...
IB Biology HL1 Fall MC questions Water / Characteristics of life
... B. Translation C. Replication D. Decomposition 33. The statement “DNA replicates by a semiconservative mechanism” means that A. Only one DNA strand is copied B. First one DNA strand is copied, and then the other strand is copied C. The two strands of a double helix have identical base sequences D. E ...
... B. Translation C. Replication D. Decomposition 33. The statement “DNA replicates by a semiconservative mechanism” means that A. Only one DNA strand is copied B. First one DNA strand is copied, and then the other strand is copied C. The two strands of a double helix have identical base sequences D. E ...
Document
... SignalSRP cleaving detaches enzyme and polypeptide cuts off signal synthesis peptide. resumes. ...
... SignalSRP cleaving detaches enzyme and polypeptide cuts off signal synthesis peptide. resumes. ...
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN: 2278-3008, p-ISSN:2319-7676.
... In the field of bioinformatics, homology modeling algorithm is regarded as an interesting site for any biological experiment and in other silico work planning. Homology modeling is the appropriate method to estimate structure related protein molecule and functional information. For 3D structure gene ...
... In the field of bioinformatics, homology modeling algorithm is regarded as an interesting site for any biological experiment and in other silico work planning. Homology modeling is the appropriate method to estimate structure related protein molecule and functional information. For 3D structure gene ...
Proteogenomics - The Fenyo Lab
... heavily on the quality of the protein sequence database (DB) • DBs with missing peptide sequences will fail to identify the corresponding peptides • DBs that are too large will have low sensitivity • Ideal DB is complete and small, containing all proteins in the sample and no irrelevant sequences ...
... heavily on the quality of the protein sequence database (DB) • DBs with missing peptide sequences will fail to identify the corresponding peptides • DBs that are too large will have low sensitivity • Ideal DB is complete and small, containing all proteins in the sample and no irrelevant sequences ...
CS790 – Introduction to Bioinformatics
... protein. If each residue can take only 3 positions, there are 3100 = 5 1047 possible conformations. • If it takes 10-13s to convert from 1 structure to another, exhaustive search would take 1.6 1027 years! ...
... protein. If each residue can take only 3 positions, there are 3100 = 5 1047 possible conformations. • If it takes 10-13s to convert from 1 structure to another, exhaustive search would take 1.6 1027 years! ...
CS790 – Introduction to Bioinformatics
... protein. If each residue can take only 3 positions, there are 3100 = 5 1047 possible conformations. • If it takes 10-13s to convert from 1 structure to another, exhaustive search would take 1.6 1027 years! ...
... protein. If each residue can take only 3 positions, there are 3100 = 5 1047 possible conformations. • If it takes 10-13s to convert from 1 structure to another, exhaustive search would take 1.6 1027 years! ...
An hierarchical artificial neural network system for the classification
... application with a pre-processing stage, represented by an artificial neural network, which attempts to classify proteins into either membrane or non-membrane proteins. Several applications of neural networks to the prediction of transmembrane segments or secondary structure prediction can be found ...
... application with a pre-processing stage, represented by an artificial neural network, which attempts to classify proteins into either membrane or non-membrane proteins. Several applications of neural networks to the prediction of transmembrane segments or secondary structure prediction can be found ...
FYVE-Dependent Endosomal Targeting of an Arrestin
... important in vivo and mechanistic insights have been unveiled in alternative models such as flies, zebrafish, worms and more recently fungi [5–12]. The social soil amoeba Dictyostelium discoideum is an attractive model system for use in studying the regulation of membrane trafficking events: it is a ...
... important in vivo and mechanistic insights have been unveiled in alternative models such as flies, zebrafish, worms and more recently fungi [5–12]. The social soil amoeba Dictyostelium discoideum is an attractive model system for use in studying the regulation of membrane trafficking events: it is a ...
Translasyon
... ribosomes are quite similar to prokaryotic ribosomes, reflecting their supposed prokaryotic origin • Cytoplasmic ribosomes are larger and more complex, but many of the structural and functional properties are similar • 40S subunit contains 30 proteins and 18S RNA. • 60S subunit contains 40 proteins ...
... ribosomes are quite similar to prokaryotic ribosomes, reflecting their supposed prokaryotic origin • Cytoplasmic ribosomes are larger and more complex, but many of the structural and functional properties are similar • 40S subunit contains 30 proteins and 18S RNA. • 60S subunit contains 40 proteins ...
Two-hybrid screening
Two-hybrid screening (also known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively.The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the binding domain (BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay.