enzymes
... The allosteric site • The allosteric site is not at the active site or substrate binding site, but is somewhere else on the molecule • The allosteric site is the site where small molecules bind and affect a change in the active site or the substrate binding site • The binding of this specific molecu ...
... The allosteric site • The allosteric site is not at the active site or substrate binding site, but is somewhere else on the molecule • The allosteric site is the site where small molecules bind and affect a change in the active site or the substrate binding site • The binding of this specific molecu ...
The Structure and Topology of Protein Serine/Threonine
... by four invariant aspartate residues and a non-conserved Glu residue (Figure 86.2b) [19]. These residues are situated at the top of the central -sandwich that forms a shallow channel suitable for the dephosphorylation of phosphoserine- and phosphothreonine-containing proteins. Six water molecules c ...
... by four invariant aspartate residues and a non-conserved Glu residue (Figure 86.2b) [19]. These residues are situated at the top of the central -sandwich that forms a shallow channel suitable for the dephosphorylation of phosphoserine- and phosphothreonine-containing proteins. Six water molecules c ...
Ch 18 reading guide
... 8. In the process, FAD is reduced to _____________ as lipoamide is reoxidized. In turn, _______________ is reoxidized to FAD as NAD+ is reduced to ________________. 9. Which cofactor also serves as a flexible linkage to bring the substrate to all the enzyme active sites? 10. Looking at Figure 18.7, ...
... 8. In the process, FAD is reduced to _____________ as lipoamide is reoxidized. In turn, _______________ is reoxidized to FAD as NAD+ is reduced to ________________. 9. Which cofactor also serves as a flexible linkage to bring the substrate to all the enzyme active sites? 10. Looking at Figure 18.7, ...
17 The Citric Acid Cycle: The latabolism of Acetyl
... the final common pathway for the oxidation of carihydrate, lipids, and protein, since glucose, fatty Is, and many amino acids are all metabolized to tylCoA or intermediates of the cycle. It also plays |major role in gluconeogenesis, transamination, mination, and lipogenesis. While several of these : ...
... the final common pathway for the oxidation of carihydrate, lipids, and protein, since glucose, fatty Is, and many amino acids are all metabolized to tylCoA or intermediates of the cycle. It also plays |major role in gluconeogenesis, transamination, mination, and lipogenesis. While several of these : ...
Document
... o average energy (dashed line) of substrates (higher on graph) and products (lower on graph) o delta G starts at substrate line and goes to transition state o adding a catalyst lowers the free energy of the transition state o In an uncatalyzed reaction, you must go through some sort of transition st ...
... o average energy (dashed line) of substrates (higher on graph) and products (lower on graph) o delta G starts at substrate line and goes to transition state o adding a catalyst lowers the free energy of the transition state o In an uncatalyzed reaction, you must go through some sort of transition st ...
O - Batavia CSD
... Induced fit model • More accurate model of enzyme action – 3-D structure of enzyme fits substrate – substrate binding cause enzyme to change shape leading to a tighter fit • “conformational change” • bring chemical groups in position to catalyze reaction ...
... Induced fit model • More accurate model of enzyme action – 3-D structure of enzyme fits substrate – substrate binding cause enzyme to change shape leading to a tighter fit • “conformational change” • bring chemical groups in position to catalyze reaction ...
No Slide Title - Docenti.unina
... surfaces. When a protein folds, exposed hydrophobic side chains get buried, and release water of its sad duty to sit against the hydrophobic surfaces of these side chains. Water is very happy in bulk water because there it has on average 3.6 H-bonds and about six degrees of freedom. So, whenever we ...
... surfaces. When a protein folds, exposed hydrophobic side chains get buried, and release water of its sad duty to sit against the hydrophobic surfaces of these side chains. Water is very happy in bulk water because there it has on average 3.6 H-bonds and about six degrees of freedom. So, whenever we ...
Name
... c. May have evolved from gibbons but not rats d. Is more closely related to humans than to rats e. May have evolved from rats but not from humans and gibbons 8. Proteins like hemoglobin and insulin have different structures because they have different ______________________, which is also known as t ...
... c. May have evolved from gibbons but not rats d. Is more closely related to humans than to rats e. May have evolved from rats but not from humans and gibbons 8. Proteins like hemoglobin and insulin have different structures because they have different ______________________, which is also known as t ...
Chapter 8 - Trimble County Schools
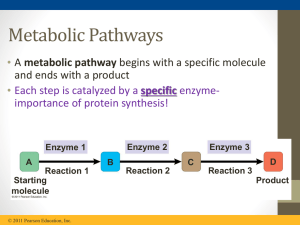
... • In feedback inhibition, the end product of a metabolic pathway shuts down the pathway • Feedback inhibition prevents a cell from wasting chemical resources by synthesizing more product than is needed ...
... • In feedback inhibition, the end product of a metabolic pathway shuts down the pathway • Feedback inhibition prevents a cell from wasting chemical resources by synthesizing more product than is needed ...
Outline 19.1 Catalysis by Enzymes
... polypeptide enters the active site (b) Hydrogen transfer allows formation of a strained intermediate (c) The peptide bond is broken. ...
... polypeptide enters the active site (b) Hydrogen transfer allows formation of a strained intermediate (c) The peptide bond is broken. ...
Question 1
... i) It should still bind because the lys is also (+) charged and can therefore still make an ionic with the (-) of the phosphate. ii) and iii) Determine if your substitution changes the ability of the enzyme and substrate to form a hydrogen bond. If it does, is that interaction now stronger or weaker ...
... i) It should still bind because the lys is also (+) charged and can therefore still make an ionic with the (-) of the phosphate. ii) and iii) Determine if your substitution changes the ability of the enzyme and substrate to form a hydrogen bond. If it does, is that interaction now stronger or weaker ...
Chemistry Option B: Human Biochemistry
... folds itself / way in which sequence is kept together by hydrogen bonding between atoms in sequence ...
... folds itself / way in which sequence is kept together by hydrogen bonding between atoms in sequence ...
462a Reading and Homework Assignment 3
... (4) Both cis and trans peptide bonds gain about 85 kJ/mol resonance energy when planar (through orbital alignment). Why are cis peptide bonds rarely seen in proteins? Why are cis peptide bonds more common for prolines than for other amino acids? Steric clash limits cis peptide bonds in most amino ...
... (4) Both cis and trans peptide bonds gain about 85 kJ/mol resonance energy when planar (through orbital alignment). Why are cis peptide bonds rarely seen in proteins? Why are cis peptide bonds more common for prolines than for other amino acids? Steric clash limits cis peptide bonds in most amino ...
Organic Chemistry Powerpoint for Bio. I
... Chemical reactions will not happen in living things without enzymes because we can’t produce enough energy available to get them to happen! Enzymes lower the activation energy of a chemical reaction so that it can happen at body temperature. This makes enzymes catalysts because they speed up chemica ...
... Chemical reactions will not happen in living things without enzymes because we can’t produce enough energy available to get them to happen! Enzymes lower the activation energy of a chemical reaction so that it can happen at body temperature. This makes enzymes catalysts because they speed up chemica ...
(a) (b)
... the cycloxygenase-2 (COX-2) enzyme. High substrate concentrations reduce the efficacy of inhibition by these drugs. These drugs are ...
... the cycloxygenase-2 (COX-2) enzyme. High substrate concentrations reduce the efficacy of inhibition by these drugs. These drugs are ...
Document
... • If a molecule can bind to another site on the enzyme (besides active site) and stop enzyme function, it is an allosteric inhibitor • Can disrupt the 3D shape of enzyme molecule so active site cannot accept substrate • Can be reversible or irreversible ...
... • If a molecule can bind to another site on the enzyme (besides active site) and stop enzyme function, it is an allosteric inhibitor • Can disrupt the 3D shape of enzyme molecule so active site cannot accept substrate • Can be reversible or irreversible ...
LECT23 Enz1
... 2. Substrate: The target of the enzyme’s action. The molecule that will undergo chemical change as a result of the enzyme 3. Enzyme activity: A measure of the enzymes catalytic effectiveness as manifested by the rate of the reaction catalyzed. 4. Cofactor: A component that works with the enzyme in e ...
... 2. Substrate: The target of the enzyme’s action. The molecule that will undergo chemical change as a result of the enzyme 3. Enzyme activity: A measure of the enzymes catalytic effectiveness as manifested by the rate of the reaction catalyzed. 4. Cofactor: A component that works with the enzyme in e ...
Purification and Characterization of Two Thermostable Proteases
... [40] purified a thiol protease from T. lanuginosus. Its molecular mass is 23.7 kDa. Hasnain et al. [17] purified a protease from T. lanuginosus. The proteinase is a thiolcontaining serine proteinase with a molecular mass of 38 kDa. Partial characteristics of proteases from these fungi are listed in ...
... [40] purified a thiol protease from T. lanuginosus. Its molecular mass is 23.7 kDa. Hasnain et al. [17] purified a protease from T. lanuginosus. The proteinase is a thiolcontaining serine proteinase with a molecular mass of 38 kDa. Partial characteristics of proteases from these fungi are listed in ...
Structural Genomics - University of Houston
... pK1 and pK2 respectively pKR is for R group pK’s pK1 2.2 while pK2 9.4 ...
... pK1 and pK2 respectively pKR is for R group pK’s pK1 2.2 while pK2 9.4 ...
Catalytic triad

A catalytic triad refers to the three amino acid residues that function together at the centre of the active site of some hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An Acid-Base-Nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine. Because enzymes fold into complex three-dimensional structures, the residues of a catalytic triad can be far from each other along the amino-acid sequence (primary structure), however, they are brought close together in the final fold.As well as divergent evolution of function (and even the triad's nucleophile), catalytic triads show some of the best examples of convergent evolution. Chemical constraints on catalysis have led to the same catalytic solution independently evolving in at least 23 separate superfamilies. Their mechanism of action is consequently one of the best studied in biochemistry.