* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Vectors in gene therapy wikipedia , lookup
Proteolysis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Lipid signaling wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
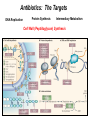
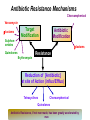
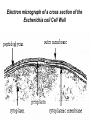
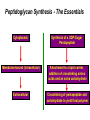
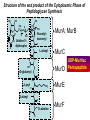
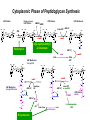
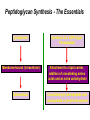
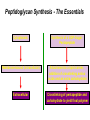
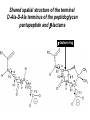
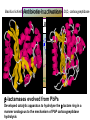
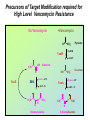
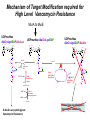
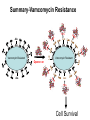
Life Death and hydrogen bonds: Bacterial peptidoglycan biosynthesis and its relationship to antibiotic resistance and the development of new antibacterials David I Roper School of Life Sciences www.warwick.ac.uk/go/ropergroup Bacterial pathogens with multiple antibiotic resistance phenotypes Staphylococcus aureus Enterococcus faecalis Klebsiella pneumoniae Pseudomonas aeruginosa Mycobacterium tuberculosis Acinetobacter spp. http://www.denniskunkel.com (2007) Antibiotic Resistance: Problem? What Problem? Virtually all prescribed antibiotics were identified between 1940-1960. Their success led to the following testimony toBut, Congress: two years earlier the first cases of penicillin-resistance in clinical isolates of Streptococcus pneumoniae were reported. In the US, penicillin resistance is currently encountered in “The United States is ready to close the book on >30% of pneumococcal infections infectious disease andDis. shift its resources to new (Doern et al 1999, Emerg. Infect. 5, 757) dimensions of health, such as chronic diseases” The US Surgeon General, Washington, 1969 Centre for Disease Control, Al. USA (Institute of Medicine (1992) Emerging infections – Microbial threats to health in the United States. National Academy Press, Washington, DC). Antibiotic Resistance Mechanisms: Nature or nuture? The problem of resistance is promoted by a number of factors: Natural Antibiotic Resistance amongst 480 Streptomyces Strains Isolated from Soil Most current antimicrobials are derived from natural sources wherein a resistance mechanism is necessary to protect the producing organism: D’Costa et al. (2006) Science 311, 374-377 Antibiotic Resistance Mechanisms: Nature or nuture? The problem of resistance is promoted by a number of factors: Most current antimicrobials are derived from natural sources wherein a resistance mechanism is necessary to protect the producing organism, but can be spread by gene transfer particularly under conditions where there is positive selection Thus, with natural product antibiotics (or derivatives thereof) The question of resistance is not if, but when……..Thanks to the medical profession and agricultural industry, this is Sooner rather than later •over prescription/use of antibiotics •Clinical environments that have allowed the spread of e.g. vancomycin resistance from Enterococcus sp. to clinicalStaphylococcus aureus strains to create VRSA •Agricultural use of antibiotics as growth promoters Denmark, 1994 24 TONS of a vancomycin derivative used for animal health – 1000X more than was used to treat human infections that year Pigs analysed for vancomycin resistant Enterococci Contained the same resistance genes as those isolated from human patients with VRE infections Dainish Government banned use of vancomycin derivatives in animal feed Antibiotics: The Targets DNA Replication Protein Synthesis Intermediary Metabolism Cell Wall (Peptidoglycan) Synthesis Antibiotic Resistance Mechanisms Choroamphenicol Vancomycin b-lactams Target Modification Antibiotic Modification Sulphonomides b-lactams Quinolones Erythromycin Resistance Reduction of [Antibiotic] at site of Action (influx/Efflux) Tetracyclines Choroamphenicol Quinolones Antibiotic Resistance, if not man made, has been greatly accelerated by man Contents and Aims To understand the action of cell wall directed antibiotics and mechanisms of resistance to them, we need: 1) A working knowledge of peptidoglycan biosynthesis: Part 1 2) An appreciation of how this process is targeted by antibiotics such as the blactams and vancomycin and a knowledge of mechanisms of resistance that have allowed pathogenic bacteria to evade the bacteriacidal effects of these cell-wall directed antibiotics: Part 2 Part 1 Peptidoglycan: Structure, Function, Synthesis and Target Peptidoglycan position in Gram-positive and Gram-negative bacteria OM PG PG CM CM Gram-positive Gram-negative Electron micrograph of a cross section of the Escherichia coli Cell Wall The Essential Role of the Peptidoglycan A Scaffold providing: Supporting and protective mesh surrounding and protecting the cytoplasmic membrane from physical forces such as osmotic pressure An anchoring point for those components of the bacterium that interact with its environment (which could be you….): extracellular proteins; Techioic Acids, Gram Positive Organisms mycolylarabinogalactan (Capsule of Mycobacterium tuberculosis) Peptidoglycan synthesis is Unique to and essential for bacterial cell viability The peptidoglycan synthesising enzymes are therefore good targets for antibiotics (both natural and man-made) NAM = N-acetyl muramic acid NAG = N-acetyl glucosamine Common examples of bacterial peptidoglycan structure Normal S.pneumoniae direct cross-link: Indirect cross-links found in penicillin-resistant Streptococcus pneumoniae: __________________________________ MurNAc GlcNAc MurNAc L-Ala 1 L-Ala D-Gln 2 D-Gln 3 L-Lys MurNAc -GlcNAc MurNAc L-Ala 1 L-Ala D-Gln L-Lys 2 D-Gln L-Lys D-Ala 3 L-Lys L-Ala L-Ala D-Ala 4 D-Ala 4 D-Ala 5 D-Ala 5 D-Ala Normal Escherichia coli and Bacillus subtilis direct cross-link Indirect cross-links found in penicillin-resistant Staphylococcus aureus ________________________________ MurNAc -GlcNAcMurNAc GlcNAc MurNAc L-Ala MurNAc L-Ala 1 L-Ala D-Gln 1 L-Ala D-Glu 2 D-Gln L-Lys 2 D-Glu DAP 3 L-Lys- Gly-Gly-Gly-Gly-Gly-D-Ala 3 DAP D-Ala 4 D-Ala 4 D-Ala 5 D-Ala 5 D-Ala GlcNac = N-acetyl-glucosamine; MurNac = N-acetyl muramic acid Peptidoglycan Synthesis - The Essentials Cytoplasmic Synthesis of a UDP-Sugar Pentapeptide Membrane-bound (intracellular) Attachment to a lipid carrier, addition of crosslinking amino acids and an extra carbohydrate Extracellular Crosslinking of pentapeptide and carbohydrate to yield final polymer Schematic Representation of the Cytoplasmic Phase of Peptidoglycan Synthesis NADP+ UDP-MurNac ATP murC ADP ATP murD ADP ATP murE NADPH murB ADP PEP murA D-Glu meso diaminopimelic acid (DAP) or lysine) OH OH O O murF UDP-GlcNac L-Ala ADP ATP Pi D-Ala-D-Ala UDP-MurNAc-pentapeptide NH 2 NH 2 OH H 2N O Gram Negative (and a few positive) H 2N Gram Positive Structure of the end product of the Cytoplasmic Phase of Peptidoglycan Synthesis OH HO N O O NH O OH O HO O O O O P P O OH OH N O O Uridine 5’diphospho N-acetylmuramyl MurA; MurB O HN L-alanyl O MurC NH O MurD HO g-D-glutamyl O HN L-Lysyl NH2 O MurE NH D-alanyl O HN D-alanine HO O MurF UDP-MurNac Pentapeptide Cytoplasmic Phase of Peptidoglycan Synthesis UDP GlcNac H O N Enoyl pyruvoyl UDP GlcNac O H O O O H H O O O O O O P P O N O O H H O P E P O H O H N m urA O H N UDP MurNac N AD P H O O + N AD P H O O H H O O O O O O O H P P N O O O H O N N O H O m urB O O H H O O O O O O P P O N O O H O AD P+P i H O O H O H N O m urC N O O H AD P+P i m urD D -G lu ATP L-G lu N O H O O O H UDP MurNacAla GluLys/DAP H O H O O O O O O O H P PO N O O H O N O H O O N O H O AD P+P i D-A la-D-A La, A T P O H N O O H N N H O O H O N H2 N H2 O AD P+P i O D-Cycloserine H O O H A T P H O D A PF UDP MurNac AlaGlu ddl H N D-A la+D-A la L-A la A lanineRacem ase N H O H O O H N O O L-lys, ATP AD P+P o rm esoD AP i H O H N N H O O O O O O N H O H O O O H H O O O O PO P O O O H N m urE H N O m urF O UDP MurNacAla GluLys/DAPAlaAla N O H N O O H O O H H O O O O O O O H P P O N O O H O H N O O H N O m urI H O O H O H O 4-[(2-napthyl)methyl] -D-Glutamate O O H H O O O O O O P P N O O O H N O O H O Fosfomycin L-Ala, ATP O O O UDP MurNacAla L,L-D iam inopim elate (D AP ) O Peptidoglycan Synthesis - The Essentials Cytoplasmic Synthesis of a UDP-Sugar Pentapeptide Membrane-bound (intracellular) Attachment to a lipid carrier, addition of crosslinking amino acids and an extra carbohydrate Extracellular Crosslinking of pentapeptide and carbohydrate to yield final polymer Membrane bound intracellular Steps of Peptidoglycan Synthesis (Streptococcus pneumonaie example) (Lipid I-Lys) (Lipid II-Lys) Lipid 2 Lipid 1 P undecaprenyl phosphate COOH COOH D-Ala NH O D-Ala HN CYTOPLASM O UMP UDP-MurNAcpentapeptide L-Lys MraY O D-Glu P COOH MurG UDPGlcNAc L-Lys Undecaprenyl D-Glu Phosphate L-Ala MurNAc NH H2 N D-Ala D-Ala P P H2 N D-Ala O D-Ala NH D-Ala MurM D-Ala D-Ala COOH HN CYTOPLASMIC FACE OFO THE L-Ala CELL MEMBRANE O L-Ala NH NH O O O HN HO O O P P O O OH O O HO OH MurNac D-Ala D-Ala Ser-tRNA Ala-tRNA D-Ala L-Lys Ser-Ala L-Lys Ser Undecaprenyl D-Glu D-Glu O Phosphate L-Lys L-Ala L-Ala NH MurNAc GlcNAc MurNAc GlcNAc MurNAc GlcNAc O P P P D-GluP P P L-LysHN D-Glu L-Ala HN O MurN GlcNac O O HN O HO HN O HO HO O O O P P O O OH O O HO OH MurNac Peptidoglycan Synthesis - The Essentials Cytoplasmic Synthesis of a UDP-Sugar Pentapeptide Membrane-bound (intracellular) Attachment to a lipid carrier, addition of crosslinking amino acids and an extra carbohydrate Extracellular Crosslinking of pentapeptide and carbohydrate to yield final polymer Membrane bound extracellular Steps of Peptidoglycan Synthesis (Example: Streptococcus pneumoniae: Ser-Ala) CELL MEMBRANE, CELL MEMBRANE, EXTRACELLULAR EXTRACELLULAR FACE FACE P P P P P P Penicillinbinding binding Penicillin protein protein carboxy-peptidase transpeptidase P P P P P P P P MurNAc GlcNAc MurNAc GlcNAc MurNAc GlcNAc MurNAc GlcNAc MurNAc GlcNAc MurNAc GlcNAc transL-Ala L-Ala L-Ala transL-Ala L-Ala L-Ala glycosylase D-Glu D-Glu D-Glu glycosylase D-Glu D-Glu D-Glu L-Lys Ser-Ala L-Lys Ser-Ala L-Lys Ser-Ala L-Lys Ser-Ala L-Lys Ser-Ala L-Lys Ser-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala EXTRACELLULAR EXTRACELLULAR SPACE SPACE D-Ala D-Ala -MurNAcGlcNAc GlcNAcMurNAc MurNAc GlcNAcGlcNAc-MurNAc L-Ala L-Ala L-Ala L-Ala D-Glu D-Glu D-Glu D-Glu L-Lys Ser-Ala L-LysSer-Ala Ser-Ala L-Lys L-Lys D-Ala Ser-Ala D-Ala D-Ala D-Ala D-Ala D-Ala Penicillinbinding binding Penicillin protein protein Trans-peptidase carboxy-peptidase D-Ala D-Ala -MurNAcGlcNAc GlcNAcMurNAc MurNAc GlcNAcGlcNAc-MurNAc L-Ala L-Ala L-Ala L-Ala D-Glu D-Glu D-Glu D-Glu L-Lys L-Lys Ser-Ala L-Lys Ser-Ala L-Lys Ser-Ala Ser-Ala D-Ala D-Ala D-Ala D-Ala The antibiotic targets of the lipid-linked steps of peptidoglycan synthesis tunicamycin, mureidomycin A, liposidomycin B Lipid-linked steps of peptid oglycan assembly UDP-Mur NAcpentapept ide D-Ala D-Ala L-Lys D-Glu L-Ala UMP MraY CYT OPLA SM ramoplanin MurNAc P P CELL SURFACE amphomycin P undecapre nyl phosphate UDPGlcNAc P P P P MurG bacitracin peptidogly can cross-linking D-Ala D-Ala L-Lys D-Glu L-Ala MurNAc GlcNAc P P P P MurNAc GlcNAc MurNAc GlcNAc L-Ala L-Ala D-Glu D-Glu L-Lys Ala-Ser L-Lys Ala-Ser Ser-Ala Ser-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala L-Lys Ala-Ser Ser-Ala D-Glu L-Ala MurNAc GlcNAc P P MurM/N Ala-tRNA Ser-tRNA P P MurNAc GlcNAc transglycosylase L-Ala D-Glu L-Lys Ala-Ser Ser-Ala D-Ala D-Ala moenomycin penicillins (b-lactams) vancomycin (glycopeptides) Summary 1) Peptidoglycan synthesis is a three phase process 2) The first cytoplasmic phase forms a UDP-sugar linked pentapeptide precursor 3) The second phase on the cytoplasmic face of the cell membrane forms a lipidsugar linked pentapeptide precursor 4) The third phase on the extracellular face of the cell membrane polymerises the lipid sugar to form the peptidoglycan 5) All phases are subject to the action of one or more antibiotics, however, clinically, the most exploited antibiotics target the third phase of peptidoglycan synthesis. Part 2 Mechanisms of Action of and resistance to Cell-Wall Directed Antibiotics 1) The b-lactams H H N Ph O OCH3 OCH3 H S N H H N O O CO2H Penicillin G H S N O CO2H Methicillin Membrane bound Extracellular Steps of Peptidoglycan Synthesis Staphylococcus aureus Enterococcus faecicum Streptococcus pneumoniae EXTRACELLULAR FACE CELL MEMBRANE, MEMBRANE, EXTRACELLULAR FACE CELL P P P P P P Penicillin binding binding Penicillin protein protein transpeptidase transpeptidase P P P P MurNAc GlcNAc GlcNAc MurNAc MurNAc GlcNAc GlcNAc MurNAc transL-Ala L-Ala transL-Ala L-Ala glycosylase D-Glu D-Glu glycosylase D-Glu D-Glu D-Asn D-Asn (Gly) (Gly) L-Lys Ser-Ala L-Lys Ser-Ala 5 5 L-Lys L-Lys D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala D-Ala EXTRACELLULAR EXTRACELLULAR SPACE SPACE D-Ala D-Ala -MurNAc GlcNAc GlcNAc -MurNAc L-Ala L-Ala D-Glu D-Glu L-Lys L-Lys D-Asn (Gly)5 D-Ala Ser-Ala D-Ala D-Ala D-Ala MurNAc GlcNAcGlcNAcMurNAc L-Ala L-Ala D-Glu D-Glu L-Lys (Gly) 5 L-Lys Ser-Ala D-Ala D-Ala P P P P MurNAc GlcNAc GlcNAc MurNAc L-Ala L-Ala D-Glu D-Glu (Gly) D-Asn L-Lys Ser-Ala 5 L-Lys D-Ala D-Ala D-Ala D-Ala Penicillin binding binding Penicillin protein protein carboxy-peptidase carboxy-peptidase D-Ala D-Ala -MurNAc GlcNAc GlcNAc -MurNAc L-Ala L-Ala D-Glu D-Glu L-Lys L-Lys (Gly)5 D-Asn Ser-Ala D-Ala D-Ala MurNAc GlcNAcGlcNAcMurNAc L-Ala L-Ala D-Glu D-Glu L-Lys Ser-Ala (Gly)5 L-Lys D-Ala D-Ala PbPs are Multimodular and Multifunctional enzymes Class A Class B S. aureus MecA (PbP2a): Monofunctional Transpeptidase (TP) TP Unknown N-terminal function Linker S. aureus PbP2 bifunctional Transpeptidase (TP)/Transglycosylase (TG) TP Linker S398 TG E171;E114 S403 Cytoplasm N Cytoplasm N Penicillin Binding Proteins (PBPs) A group of transpeptidases (class A & B) and d,d carboxypeptidases (class C) that utilise a serine active site nucleophile: MurNAc GlcNAc - MurNAc GlcNAc MurNAc GlcNAc 1 2 3 4 - O 2C L-Ala D-Glu L-Lys D-Ala NH 5 D-Ala O 1 2 3 4 O H NH3+ 5 NH L-Ala Transpeptidase D-Ala L-Lys D-Ala H2N O MurNAc GlcNAc NH3+ Ser Lys O 4 L-Lys 3 D-Glu 2 L-Ala 1 D-Glu O 5 O2 C 1 L-Ala 2 D-Glu 3 L-Lys 4 D-Ala O H MurNAc GlcNAc 1 L-Ala 2 D-Glu 3 L-Lys 4 D-Ala CO2- H tetrapeptide sidechain D-Ala N H D-Ala L-Lys D-Glu L-Ala NH3+ Lys D,D-carboxypeptidase Lys O Ser H Ser O cross-linked peptidoglycan 5 4 3 2 1 MurNAc GlcNAc b-Lactam Antibiotics 65% world market of antibiotics >50 marketed drugs of this class H H Penicillins N Cephalosporins Ph Carbapenems O Monobactams Cephalosporin-penicillin hybrids, O Penems H S N CO2H Strained, reactive b-lactam ring Shared spatial structure of the terminal D-Ala-D-Ala terminus of the peptidoglycan pentapeptide and b-lactams b-lactam ring b-Lactams Antimicrobial Suicide Substrates Antimicrobial Potency arises because the drug simultaneously targets mutiple enzymes in peptidoglycan synthesis (7 PbPs in E. coli, 5 in S. pneumoniae) Antimicrobial Potency arises because the drug exploits its strained b-lactam ring structure and the catalytic apparatus of the PbP to spring a trap on the unsuspecting enzyme…. H H H N COR H S H3C H3C N O O - O2C H H3C H3C - NH3+ H S H N COR O NH O2C O NH2 Ser Ser Lys Lys PBP's irreversible acylation (or slow hydrolysis) RESULT: Inhibition of peptidoglycan crosslinking leading to a weakened cell wall, leading to osmotic rupture of the cell membrane and cell death Emergence of penicillin resistance Antibiotic Inactivation b-lactamases Principally Gram negative enteric and Pseudomonad pathogens, exception: Staphylococcus aureus Target Modification PbP Remodelling Principally Gram positive pathogens, e.g. Streptococcus pneumoniae PbP Re-aquisition Principally Gram positive pathogens, e.g. MRSA, PbP2a Bacillus lichiniformis b-lactamaseInactivation Streptomyces D,D, carboxypeptidase Antibiotic b-Lactamases- Like PbPs but not H H H N COR S H3C H3C - N O2C O O H H3C H3C S H H HN COR O NH O - O2C NH3+ H2O NH2 Ser H3C b-lactamases H3C Ser Lys Lys S H H HN COR CO2- NH OH - O2C NH3 + Ser Lys PBP's irreversible acylation (or slow hydrolysis) b-lactamases evolved from PbPs Developed catalytic apparatus to hydrolyse the b-lactam ring in a manner analogous to the mechanism of PbP carboxypeptidase hydrolysis Target Modification Global clonal spread of penicillin resistant pneumococci 1984‘86 ‘88 ‘90 ‘92 ‘94 ‘96 ‘98 2000 Serotype 23F Spain UK France South Korea USA South Africa Hungary Iceland Bulgaria Portugal Germany Thailand Colombia The Netherlands Argentina Denmark Japan Malaysia Singapore Taiwan Mosaic Gene Structure In Pneumococcal pbp2x generated from homologous recombination with homologues from closely related Streptococci Transpeptidase Domain Penicillin sensitive strains (mic 0.02 mg/ml) pbp2x Ser A B Penicillin resistant strains C (mic ≤16 mg/ml) D E F Generation of a penicillin-resistant pneumococcal PbP2x by homologous recombination K-[T/S]-G K-[T/S]-G S-X-X-K S-X-X-K 341 341 A337 Thr337 338338 [S/Y]-X-[N/C] [S/Y]-X-[N/C] Generalised active of site a PbP with aminoacids acids from from penicillin Generalised active site scaffold a of PbP with amino penicillin Sensitive PbP2x from S. pneumoniae R6 superimposed upon it Resistant PbP2x from S. pneumoniae Sp328 superimposed upon it PbP2x crystal structure reveals penicillin resistance by target modification has a cost •PbP2x sequences with up to 20% divergence between resistant and sensitive strains, aquired through homologous recombination •Key mutations distort the transpeptidase active site •Optimal distances between conserved active site residues changed, causing simultaneous loss of catalytic activity (to 1 thirtieth of rate shown by sensitive PbP2x) and aquisition penicillin resistance. •Implied consequence is that penicillin resistance exacts a price on cell wall synthesis, whose rate of cross linking will be impaired Mechanisms of Action of, and resistance to Cell-Wall Directed Antibiotics 2) Vancomycin and other Glycopeptides O Me OH NH2 HO HOCH2 Me O O O HO O H N H OH O H N H -O C 2 Cl Cl HO O O H H H N O O H N O H N H H O NH2 HO OH HO Vancomycin N H O Me NH+ 2 Vancomycin - A Vital Antibiotic • Vancomycin is the last line of defence against Gram-positive bacteria where other treatments fail, Staphylococci. Streptococci, Corynebacteria, Clostridia and particularly MRSA. • Contraindications Deafness, Severe hypertension (red man syndrome), nausea, diarrhoea, vomiting, may lead to other fungal and gram-negatives. • Glycopeptide resistant Enterococci (GRE) known since the late 1980s Some GREs are untreatable due to multiple antibiotic resistance mechanisms. Vancomycin : Mode of Action s UDP-Mur NAcpentapept ide UMP D-Ala D-Ala L-Lys D-Glu L-Ala D-Ala D-Ala L-Lys D-Glu L-Ala MurNAc GlcNAc P P MurG •Vancomycin is not an enzyme inhibitor. MraY PLA SM MurNAc UDPGlcNAc MurM/N Ala-tRNA Ser-tRNA D-Ala D-Ala L-Lys Ala-Ser D-Glu L-Ala MurNAc GlcNAc P P •Vancomycin bindsPP to the D-alanyl-D-alanine termini of peptidoglycan P units prior their incorporation in the cell wall: P P P URFACE Extracellular surface yl peptidogly can cross-linking P P MurNAc GlcNAc MurNAc GlcNAc L-Ala L-Ala D-Glu D-Glu L-Lys Ala-Ser L-Lys Ala-Ser D-Ala D-Ala D-Ala D-Ala P P MurNAc GlcNAc transglycosylase L-Ala D-Glu L-Lys Ala-Ser D-Ala D-Ala Vancomycin (glycopeptides) •By doing so, it prevents transpeptidation reactions from crosslinking adjacent peptidoglycan chains, weakening the cell wall leading to osmotic stress and lysis Vancomycin: Targets the D-Ala-D-Ala Terminus Extracellular Peptidoglycan Precursors VANCOMYCIN Me O OH NH2 HO HOCH2 Me O O O HO O Cl HO H -O C 2 H N H OH O H N O H H N H O O H N H O N H H O NH2 HO OH HO R Mr=1805 Cl O H Me O N H O- H N O O H Me N-Ac-D-Ala-D-Ala N H O Me NH+ 2 Emergence of Vancomycin Resistance Reduction of [Antibiotic] at site of Action Target Modification “Visa”: Peptidoglycan Remodelling Vancomycin-intermediate resistant Staphylococcus aureus – resistance by decreased permeability using a thicker peptidoglycan layer mic: ≥16 mg/ml (Sensitive: 0.02 mg/ml) Principally Gram positive pathogens,e.g. Enterococci and more recently (2002) Staphylococcus aureus (“VRSA”) mic: ≥500 mg/ml Target Modification mediated Mechanisms of Vancomycin Resistance VANCOMYCIN O VANCOMYCIN OH Me O OH Me NH2 NH2 HO HOCH2 H O O H N H H H N H H N H O N H H N H O NH2 OH HO R H Me O N H H N NH+ -O C 2 H H N H H H N H H O H Me N-Ac-D-Ala-D-Ala Vancomycin sensitive H N H N H H Me NH2+ O O NH2 OH HO O- O O N HO O O Me 2 O HO O OH O H N O O Cl O Cl HO OH O H N -O C 2 Cl Cl HO O HO O Me O O O HO O HO HOCH2 Me O O R O- H Me O O N H O O H Me N-Ac-D-Ala-D-Lactate Vancomycin Resistant 1000-fold drop in affinity of vancomycin for its target Vancomycin Resistance; Simple and elegant in principle, Loss of a single hydrogen bonding interaction by interconverting DAlanine to D-lactate at the end of the peptidoglycan peptide eliminates the interaction of vancomycin with its target ……..complex in execution In Gram positive pathogens such as Enterococcus faecalis and Enterococcus faecicum Vancomycin resistance is more complex than target modification mediated penicillin resistance, because modification of a single (PbP) gene can be sufficient for b-lactam resistance. Vancomycin resistance, however, requires modification of complex metabolites such as those at the end of peptidoglycan synthesis and so requires expression of many different genes involved in the synthesis of the new target. ……mechanism to spread between pathogens Transposon encoded high-level vancomycin resistance operon. Has been transferred from an Enterococcus to S. aureus in a clinical setting !!!!!!!!! Sensing and initiation of gene expression leading to Enterococcal Vancomycin Resistance ? Cell membrane VanS PO4 Cytoplasm PO4 VanR VanR PvanR vanR vanS Response regulator Sensor Regulation PvanH vanH vanA vanX D-lactate producing reductase D-Ala-D-lac D-Ala-D-Ala ligase dipeptidase Resistance Precursors of Target Modification required for High Level Vancomycin Resistance No Vancomycin +Vancomycin CO2 O Pyruvate NADPH VanH NADP+ + H3N OH D-Alanine O VanX CO2 HO D-Ala + ATP DDL D-Lactate D-Ala + ATP VanA ADP + Pi + H3N O N H CO2 D-Alanyl-alanine ADP + Pi + H3N O CO2 O D-Alanyl-D-lactate Mechanism of Target Modification required for High Level Vancomycin Resistance MurA to MurE UDPmurNac AlaGluLys/DAP/Ala-Lac HO N OH O O OH HO O O O P O P O O OH HO OH O OH N N O N O O OH HO O O O P O P O O OH O OH OH N O N O O O OH HO O O O P O P O O OH O OH N O N HN O O HN O O O m urF NH O HO O N HN O UDPmurNac AlaGluLys/DAP/AlaAla UDPmurNac AlaGluLys/DAP NH O O m urF NH O HO HO O HO O HN HO D-Ala-D-Lac, ADP + Pi ATP O O OH VanA ADP + Pi D-Ala-D-Lac Ligas e HO O NH2 O NH O O ATP D-Ala + D-Lac O D-Ala-D-ALa NH2 O HO HN HO VanX: D-Ala-D-Ala Dipe ptidas e ATP NH2 O NH D-Ala + D-Ala ADP + Pi D-Ala-D-Ala Ligas e O HN HO O Van H: D-Lactate de hydroge nas e NAD+ NADH Py ruv ate D-Ala-D-Lac peptidoglycan: Vancomycin Resistance O HN D-Ala-D-Ala peptidoglycan: Vancomycin sensitivity Summary – Vancomycin Sensitivity Ala Ala Vancomycin sensitive Ala Ala Ala Ala Vancomycin sensitive Ala Ala Cell Death Summary-Vamcomycin Resistance Ala Ala Vancomycin Resistant Ala Lac Lac Ala Vancomycin Resistant Operon on Ala Ala Ala Lac Cell Survival Overall Summary 1) Penicillin acts as a suicide substrate that chemically modifies a group of mechanistically related cell wall polymerizing/modifying enzymes: the Penicillin binding proteins 2) Penicillin resistance involves antibiotic inactivation (b-lactamases); target modification (Streptococcal PbPs) or wholesale target replacement (S. aureus MecA) 3) Vancomycin acts not on a protein but on late peptidoglycan intermediates to which it binds via their terminal D-alanyl-D-alanine residues. This sterically prevents transpeptidation of cell wall precursors leading to cell lysis. 4) Resistance to vancomycin is caused by modification of its target causing loss of a single hydrogen bond interaction, by replacment of D-alanyl-D-alanine with D-alanylD-lactate. This is relatively complex to achieve, requiring re-programming of the synthesis of the peptidoglycan. References Bacterial antibiotic resistance and discovery: 1. Walsh, C. (2000) Molecular mechanisms that confer antibacterial drug resistance, Nature 406, 775-781. 2. Gwynn, M. N., Portnoy, A., Rittenhouse, S. F., and Payne, D. J. (2010) Challenges of antibacterial discovery revisited, Ann N Y Acad Sci 1213, 5-19. 3. Agarwal, A. K., and Fishwick, C. W. (2010) Structure-based design of anti-infectives, Ann N Y Acad Sci 1213, 20-45. Vancomycin resistance 4. Healy, V. L., Lessard, I. A., Roper, D. I., Knox, J. R., and Walsh, C. T. (2000) Vancomycin resistance in enterococci: reprogramming of the D-ala-D-Ala ligases in bacterial peptidoglycan biosynthesis, Chem Biol 7, R109-119. Peptidoglycan specific A comprehensive and detailed set of reviews of many aspects of bacterial cell wall biogeneisis and inhibition. 5. Ende, J. C. a. A. V. D. (2008) Peptidoglycan: the bacterial Achilles heel, FEMS Microbiol Reviews 32, 147-408. 6. Bugg, T. D., Braddick, D., Dowson, C. G., and Roper, D. I. (2011) Bacterial cell wall assembly: still an attractive antibacterial target, Trends Biotechnol 29, 167-173. 7. Mattei, P. J., Neves, D., and Dessen, A. (2010) Bridging cell wall biosynthesis and bacterial morphogenesis, Curr Opin Struct Biol 20, 749-755.