* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Human Molecular Evolution Lecture 2
Origin of language wikipedia , lookup
Multiregional origin of modern humans wikipedia , lookup
Origins of society wikipedia , lookup
Archaic human admixture with modern humans wikipedia , lookup
Behavioral modernity wikipedia , lookup
Human genetic clustering wikipedia , lookup
Race and genetics wikipedia , lookup
Discovery of human antiquity wikipedia , lookup
Before the Dawn (book) wikipedia , lookup
Evolutionary origin of religions wikipedia , lookup
A30-Cw5-B18-DR3-DQ2 (HLA Haplotype) wikipedia , lookup
Recent African origin of modern humans wikipedia , lookup
Anatomically modern human wikipedia , lookup
Human genetic variation wikipedia , lookup
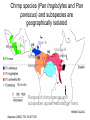
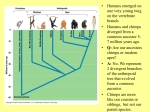
Human Molecular Evolution Lecture 1 Molecular evolution – Humans as apes You can download a copy of these slides from www.stats.ox.ac.uk/~harding We are apes! though a unique form of ape • What makes us different from other apes? Apes are primates Great apes are shown. Gibbons also are apes, (i.e. lesser apes). Homo sapiens (human) Pongo pygmaeus (orangutan) Gorilla gorilla (lowland gorilla) Pan troglodytes (common chimpanzee) Gibbon (lesser ape) Primate classification In 1735 Linneas classified humans in the same taxonomic group with other living primates: apes, monkeys, lemurs, lorises, and tarsiers, in his Anthropomorpha, now Order: Primates. Ring tailed lemur Pygmy loris Spider monkey Tarsier The beginnings of the study of primate evolution Charles Darwin • 1830s: first discoveries of primate fossils, providing evidence of a temporal dimension in primate diversity and biogeography, including extinct species & evidence that apes once lived in Europe. • Publication of Darwin’s Origin of Species (1859) and The Descent of Man and Selection in Relation to Sex (1871): explaining how evolution could take place. – ‘much light will be thrown on the origin of man and his history’ Primates: basic design • Generalised arboreal anatomy, but many examples of specialized adaptations for locomotion, e.g. – Knuckle walking by chimpanzees and gorillas, but not by humans. • Stereoscopic vision • Most have opposability of their thumbs and/or first toes to make for a grasping hand • Many have relatively large brains for their body size • Life history traits: variation in gestation length, age of weaning, age at sexual maturity (the high end of which is not found in other primates of comparable body size), that emphasize high investment in small numbers of offspring, learned behaviour and sociality. Primatology as a basis for the study of human evolution • By the beginning of the 20th century, primate evolution had become established as an area of major interest within anthropology – providing the broad evolutionary context for studying human origins. • Classification by morphological similarity is challenged by phylogenetic methods. • What was the branching order of these species? – Chromosomes (karyotypes) – Molecules (proteins and DNA) • What was the time scale? – Fossils – Molecules Tarsier Primate Phylogeny Pygmy Loris Higher Primates (including apes and monkeys) and Tarsiers are Haplorhines Divergence between Haplorhines vs Strepsirhines (bushbabies, lorises and lemurs) Note the time scales. Supported by fossils Estimated from molecular divergence Branching order and time-scale for ape phylogeny Hla: Hylobates lar (gibbon) 1 Ggo: Gorilla gorilla 2 Ptr: Pan troglodytes (common chimp) 3 Ppa: Pan paniscus (bonobo) 4 5 Hsa: Homo sapiens (humans) Ppy: Pongo pygmaeus (orangutan) Hacia JG (2001) What traits distinguish humans from other apes? • • • • • • • • • • Body shape, S-shaped spine Relative limb length Efficient bipedal locomotion Skull balanced upright on vertebral column Cranial properties, relative brain size and brain topology Small canine teeth Long ontogeny (development time) and lifespan Reduced body hair Language Advanced tool making How did these traits evolve? Evidence from hominin fossils Bipedal mode of location, evident for earliest hominins – australopithecines Large brain, disproportionately large for body size; evolved ~2 MYA, characteristic of Homo Our closest living relatives are chimps Nature 437(7055):17-19, 2005 Chimpanzees: two species (and several subspecies) Pan troglodytes (common chimp) Pan paniscus (bonobo) What can we learn from studies of chimps? Chimp species (Pan troglodytes and Pan paniscus) and subspecies are geographically isolated Niger R. Western Sanaga R. Ubangui R. Eastern Central Bonobo Ranges of chimp species and subspecies appear bounded by rivers. Gagneux (2002) TIG 18:327-330 Gorilla Bonobo P.t. schweinfurthii (Eastern) Human P.t. troglodytes (Central) P.t. verus (Western) Figure from Stone et al. (2002) Unrooted phylogenetic tree (maximum parsimony) for Y chromosome haplotypes The tree indicates that some nucleotide differences discriminate chimp subspecies. What does this impIy for FST? Reconstructing haplotypes from the tree Pp3 Pp2 Pp1 Bonobo Ptv1 Ptv2 Western Ptt1 Ptt2 Ptt4 Ptt5 Central The actual locations of variable sites are unknown, but we have some information about how variable sites are shared between haplotypes. Estimating FST from Y haplotypes FST = 0 (min value) when allele / haplotype lineages frequencies are the same across subpopulations, ie variance (s2) is zero. FST = 1 (max value) when alternative alleles / haplotype lineages are fixed (100% freq) in different subpopulations. The Y haplotype tree indicates that chimp subpopulations have diverged and do not share haplotype lineages. FST ~ 1 The Western and Central common chimpanzee (P. troglotydes) subpopulations have diverged nearly as much from each other as from P. paniscus (bonobo). MtDNA likewise discriminates chimp sub-’species’. Estimating diversity from average pairwise sequence difference Bonobo Pp3 freq= 2 Pp2 freq= 2 Pp1 freq= 4 Pp1 Pp1 Pp1 Pp1 Pp2 Pp2 Pp3 Pp3 Pp1 Pp1 Pp1 Pp1 Pp2 Pp2 Pp3 Pp3 0 0 0 0 3 3 3 3 0 0 0 0 3 3 3 3 0 0 0 0 3 3 3 3 0 0 0 0 3 3 3 3 3 3 3 3 0 0 1 1 3 3 3 3 0 0 1 1 3 3 3 3 1 1 0 0 3 3 3 3 1 1 0 0 Av = 104/64 = 1.625 Divide Av by the length of sequence to give nucleotide diversity, p. Gorilla Bonobo P.t. schweinfurthii (Eastern) Human P.t. troglodytes (Central) P.t. verus (Western) Figure from Stone et al. (2002) Unrooted phylogenetic tree (maximum parsimony) for Y chromosome haplotypes The tree indicates that Bonobo and Central chimps have similar levels of diversity, but that Western chimps have less diversity. Phylogenetic tree (maximum likelihood) of chimpanzee and bonobo Xq13.3 haplotypes B: Bonobo, Pan paniscus C: Central African, P. t. t. W: Western, P. t. verus E: Eastern, P. t. schweinfurthii Kaessmann et al. 1999 Science 286:1159-1162 Haplotypes do not completely discriminate subspecies. What does this imply for FST ? Implications for FST and p The tree shows incomplete lineage sorting for Xq13.3 haplotypes. There is a high level of population divergence but some Central chimp haplotypes are mixed in with the Western chimp haplotypes, so FST <1 Note that the average sequence difference between pairs of haplotypes (p) taken for Western chimps will be smaller than the average sequence difference between pairs of haplotypes (p) taken for Central chimps. Diversity for Central chimps is higher than for Western chimps. Comparing human and chimp genomes 15 Feb 2001 Heterozygosity, estimated by p (av. sequence difference between two chromosomes) p = 7.51 x 10-4 i.e. 1 SNP per 1,331 bp NIH diversity panel (including African American, European, Chinese) 1 Sept 2005 p = 9.5 x 10-4 for Clint (from West Africa) p = 9.5 x 10-4 among 4 West African chimps p = 17.6 x 10-4 among 3 Central African chimps. West African chimps have similar genomic diversity to humans. Central African chimps have twice as much diversity. Ne for Chimps (P. troglodytes) ? Chimpanzee MtDNA NRY (3 kb) *Stone et al. (2002) PNAS 99:43-48 Xq13.3 (10 kb) Kaessmann et al. (1999) 50 autosomal loci Yu et al. (2003) Genetics 164:15111518 % sequence differences *Nef =41,000; *Nem= 21,000 p=0.0007; qW=0.0011 Ne=35,000 p=0.0013; Ne=20,900 (P. troglodytes) Ne = 20,100 (P.t.t.) Ne = 14,600 (P.t.s.) Ne = 13,000 (P.t.v.) ~3-7 times higher in chimps than in humans for mtDNA, NRY and Xq13.3 ~1-2 times higher in chimps than in humans for autosomal loci. Differences between loci • Why are there greater sequence differences among chimps in Y haplotypes and mtDNA compared with autosomal genomes? • Isolation between chimp subpopulations leads to genetic divergence, lineage sorting and accumulation of fixed differences. This effect has added sequence differences in addition to polymorphism in Y haplotypes and mtDNA but not to autosomal loci. • The estimates of Ne for chimps from Y haplotypes and mtDNA are incorrect because they should be based only on polymorphism, not on fixed differences. What is effective population size, Ne? • An estimate of Ne from autosomal genetic diversity: N^e = p / 4.m • In the model, Ne is inversely proportional to how genetic drift has enhanced or eroded polymorphism. • In the data, p is an estimator of Nem provided that it is based on polymorphism. • Two issues for interpreting sequence diversity: – Size (Ne) – diversity shared by larger numbers of breeding individuals in a population is less subject to erosion by genetic drift. – Structure – a number of individuals in a structured population (island model) may present more sequence differences than the same number in a randomly-mating population. Comparing humans with chimps: population structure Humans • Diversity in racial phenotypes does not mark genetic divergence. • A moderate level of structure: FST ~0.15 between populations, across most classes of polymorphism, though lower than this for (microsatellites) and higher (~0.33-0.38) for Y haplotypes. • Largest genetic distances are between populations within sub-Saharan Africa not between populations on different continents Chimpanzees • Diversity in phenotypes does mark genetic divergence between bonobo and common chimps but not divergence between chimp subpopulations. • A high level of structure and isolation leading to divergence. Even higher for Y chromosomes than for autosomal loci. • Genetic distances between subpopulations almost as large as between species. Comparing humans with chimps: differential selective pressures? • Morphological diversity is low in chimps compared with humans. Is this due to strong differential selection in humans. • Classical polymorphisms (blood groups) and enzyme polymorphisms have higher diversity in humans than in chimps. • MHC diversity, for HLA-A in particular, is lower in chimps than in humans. • Levels of polymorphism at VNTRs, dinucleotide microsatellites in particular, seem reduced in chimps compared with humans. Ascertainment bias is a partial but incomplete explanation. • How have patterns of selection differed in humans compared with chimps? Local adaptations to climate? And to pathogens? Conclusions • One feasible assumption is that hominins during the Pleistocene were a highly structured species, with species and sub-species differentiation like in chimps today. • Then from contemporary levels and patterns of genetic diversity we can suggest that modern humans descend from a single regional subpopulation and reject the multiregional hypothesis. • The estimates of ~10,000 for Ne for humans and for western chimps implies that neutral diversity is being lost by genetic drift as expected in long term small populations • The estimate of 20,000 for Ne for central chimps implies a larger long term evolutionary size. References • Bramble DM and Lieberman DE (2004) Endurance running and the evolution of Homo. Nature 432: 352345. • Hacia JG (2001) Genome of the apes. Trends in Genetics 17(11): 645-637 • Gagneux P (2002) The genus Pan: population genetics of an endangered outgroup. Trends in Genetics 18:327330