* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Hologenome theory of evolution wikipedia , lookup
Natural selection wikipedia , lookup
Gene expression programming wikipedia , lookup
Evolutionary developmental biology wikipedia , lookup
The eclipse of Darwinism wikipedia , lookup
Molecular paleontology wikipedia , lookup
Saltation (biology) wikipedia , lookup
Organisms at high altitude wikipedia , lookup
Evidence of common descent wikipedia , lookup
Genetics and the Origin of Species wikipedia , lookup
Koinophilia wikipedia , lookup
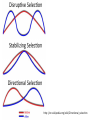
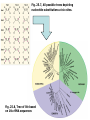
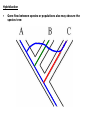
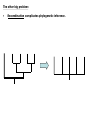
Chapter 23 - Molecular evolution: Types of questions: • • How do genomes, DNA, and protein sequences evolve? • Dynamics and mode of change. • Rates of change. How are genes and organisms evolutionarily related? • Phylogenetic systematics/trees/networks • Species concepts (allopatric/sympatric) Different time scales: • Short-term: ‘population genetics’ tends to focus on genetic changes between generations and within species or between very closely related species. • Long-term: ‘molecular systematics’ tends to focus on genetic changes over many generations; departures from Hardy-Weinberg equilibrium can become significant, leading to speciation. Some basics: Homology = refers to a structure, behavior, or other character of two taxa that is derived from the same or equivalent feature of a common ancestor. • Homology applies to nucleotide sequences: • Positional vs. character homology GTACCT G-ATCT 1. Four of six nucleotide positions have undergone no change. 2. A substitution has occurred at position 4. 3. Insertion/deletion has occurred in one sequence at position 2. Sequence alignment: • Rapid sequence divergence or divergence over many generations can leave little in common between two sequences and make alignment difficult or impossible. • Indels ~ may be impossible to distinguish between an insertion in one sequence and a deletion in another sequence. example: mtDNA 12S rRNA in six different genera CCACCT-GT---TTCAAAA-CTCAGGCCTT TCACCTAGC---TCCAAA--C-TAGGCCTT CTGCCT-AC---TTCCC---C-CAGGCCTT TCGCCT-AC---T-CAA---C-CAGGCTTT TCGCCT-ACATTTTCCC---C-CAGGCTTT • Many alignment methods exist; all use algorithms that seek to maximize the number of possible matching nucleotides or amino acids and minimize the number of indels. Jukes-Cantor (1969) model of nucleotide substitution: • Alignment of sequences with many differences underestimates the actual number of substitutions. • PC(t) = 1/4 + (3/4)e-4t = rate of substitution • # substitutions/site = K =-3/4ln(1-4/3p) p = % difference (raw count) Fig. 25.1 Saturation of DNA Sequences Transversions Transitions http://www.ccg.unam.mx/~vinuesa/images/Ti_tv_saturation_plot.png Rates of nucleotide substitution between sequences: • Rate = r = K/(2T) *2T because substitutions accumulate simultaneously and independently in both sequences (two lineages). taxon 1 taxon 2 Rates of nucleotide substitution (cont.): • Different genes evolve at different rates. • Coding regions and non-coding regions differ. • Different parts of the coding region differ: 3rd pos. are 2- and 4-fold degenerate synonymous substitution • Synonymous substitution ~5X > than non-synonymous substitution. • Substitution ≠ mutation • Substitution implies that the mutation has passed through the filter of selection. • Synonymous subsitution ~ mutation rate • Non-synonymous substitution ≠ mutation rate. Rates of nucleotide substitution (cont.): • Most substitutions in 3’-flanking regions are tolerated. • Rates: 3’ regions > introns > exons • 5’ regions < 3’ regions due to the presence of promoters and other regulatory elements. • Leader and trailer regions < 5’ regions; important for mRNA processing and translation. • Highest overall rates of substitution occur in non-functional pseudogenes and other non-functional, non-coding sequence such as microsatellites. Codon usage bias: • Some synonymous codons are favored over others e.g., yeast Leu codons: 6 possible codons/80% are UUG • Repeated evolution of the same amino acid in hemoglobin subunits of Andean ducks---all involve the same codon. Possible explanations: • All involve selection. • Some tRNAs may be more abundant or efficient; bonding energy may differ due to differences in base pairs. • Selection is expected to be more intense for genes expressed at higher levels/organisms with short generation times. • Codon usage bias permits smaller number of tRNAs (e.g., Wobble effect). Variation in evolutionary rates: Useful for evaluating differences in substitution frequency and action of natural selection on a locus. Three types of selection operate on genotypic variation: Directional selection – natural selection favors one particular genotypic variant or phenotype over others in one particular environment, causing the allele frequency to shift. Purifying selection – removal of deleterious alleles (e.g., elimination of most non-synonymous substitutions) Balancing selection – multiple alleles are selected for in the gene pool and maintained at frequencies above the mutation rate (overdominance is a type of balancing selection). http://en.wikipedia.org/wiki/Directional_selection Michael Bamshad & Stephen P. Wooding - Nature Reviews Genetics 4, 99-111 (February 2003) Examples of balancing selection: Light and dark-colored moths in the same population Major histocompatibility complex (MHC) • Genes are important in immune response and are under selective pressure to diversify (diversifying selection). • ~90% of humans receive different MHC genes from each parent. • Sample of 200 humans will have 15-30 different alleles. • Important mechanism for outcrossing in humans. • Humans select mates on the basis of their MHC compatibility. McDonald-Kreitman (1991) test: • Compare non-synonymous/synonymous ratios within species to between species (KNS/KS). • If ratios differ, selection may be responsible. • KNS/KS typically differs by a factor of 2; but may vary 1,000-fold between different types of genes. • Conservative test. Rates of evolution in mtDNA and chloroplasts: • Organelle genomes (mtDNA, cpDNA) are distinct from nuclear genomes and show increased rates of substitution. • ~10X greater than nuclear genes. • Possible explanations: • Lack proofreading • Different DNA repair mechanisms • Higher levels of oxidative mutagens due to metabolism • Lower selective pressure; most cells contain several dozen mitochondria. • Maternally inherited; smaller effective population size; increased effects of genetic drift and selective sweeps on mtDNA/cpDNA variants that are beneficial. Molecular clocks (fig. 25.3): • Zuckerkandl and Pauling (1969): recognized that genes with similar functions generally show uniform rates over long periods of time. Molecular clocks (cont.): • Can be used to estimate divergence time. • “Clocks” tick differently in different proteins. Relative rate test (Sarich and Wilson 1973): • Measure # substitutions between two taxa and an outgroup taxon that shares a common ancestor. • doutgroup-1 > dooutgroup-2 taxon 1 outgroup taxon 2 Causes of fast/slow molecular substitution rates (cont.): • Substitution rates are expected to be related to germ line replication (or generation time). • Metabolic rate also is thought to be an important factor (correlates with body size and generation time). example: rodents are small, have a high metabolic rate, and have short generation time/rodent rates are ~2x humans and apes. • In addition to variation between and among genes, rates vary widely among taxonomic groups. • Other sources of variation: • DNA repair mechanisms/efficiency • Exposure to mutagens • Opportunities to adapt to new environments, may lead to bursts of rapid evolution. How to build trees - phylogenetic systematics-concepts/definitions: Taxon Monophyletic group of organisms recognizable by a set of shared characters and sufficiently distinct from other such groups to be ranked in a taxonomic category. Category Hierarchical level to which taxa are assigned in a classification (e.g., kingdom, phylum, class, order, etc.). Monophyly Descent from a common ancestor; every true taxon is monophyletic. Polyphyly Descent from more than one ancestral lineage. Phylogenetic systematics-more concepts/definitions: Homology Shared similarity derived from common ancestry. Homoplasy Similarity derived from convergence, parallelism, or reversal. Convergence Independent acquisition of a similar character by two or more taxa whose common ancestor lacked that character; generally refers to more distantly related lineages. Ancestral lineages possessed different character states. Parallelism Independent acquisition of the same or similar characters by more closely related lineages (i.e., similar to convergence). Ancestral lineages possessed the same character state. Reversal Reappearance of an ancestral character as the result of the loss of a derived character. Phylogenetic trees: • • Branching patterns (trees) depict genealogical relationships • applies to pedigree analysis and systematics • Useful for molecular/non-molecular data The 3-taxon example: Fig. 25.5 Phylogenetic trees (cont.): # of possible rooted trees = (2n -3)!/(2n-2(n-2))! # of possible unrooted trees = (2n -5)!/(2n-3(n-3))! # taxa # rooted trees 3 3 4 15 5 105 6 945 7 10,395 8 135,135 9 2,027,025 10 34,459,425 Finding the “best” tree: • Long tradition of using characters (morphological and molecular). • Ernst Haeckel (1866) • 1950-1960s Numerical phenetics • Willi Hennig (1966) Cladistics Willi Hennig’s cladistic characters: Synapomorphy: shared derived homologous characters inferred to have been present in the nearest common ancestor of two or more taxa, but not in earlier ancestors outside this group (phylogenetically informative). Symplesiomorphy: shared ancestral homologous characters inferred to have been present in the nearest common ancestor of two or more taxa, and in earlier ancestors outside this group (phylogenetically non-informative). Autapomorphy: unique derived character present in only one of two sister groups (phylogenetically non-informative). Tree reconstruction methods: Genetic distance: • Create a matrix of genetic distances describing genetic distances between all pairs of taxa. • Select tree that minimizes total genetic distance (distances can be weight or unweighted). Parsimony: • Minimize number of steps required to evolve shared derived homologous characters (synapomorphies) on the tree (characters may be weighted or unweighted). • Shortest tree is the best tree by principle of parsimony; i.e., the explanation that requires the fewest assumptions is preferred to other more complex explanations. Maximum likelihood/Bayesian methods: • Similar character based approaches but use ‘statistical’ methods such as maximum likelihood and attempts to model DNA evolution as we know it (assuming different frequencies of nucleotides, substitution rates, etc.). Most parsimonious taxon 1 taxon 2 outgroup taxon 1 outgroup Less parsimonious taxon 2 Fig. 25.7, All possible trees depicting nucleotide substitutions at six sites. Fig. 25.8, Tree of life based on 16s rRNA sequences Fig. 25.2, mtDNA lineage relationships in pocket gophers. Problems with tree reconstruction methods (cont.): • One or more (perhaps many) trees may best describe the data. • Equally parsimonious/likely trees may not be consistent (character support can be assed in different ways; e.g., bootstrap resampling). • Gene trees and species trees: a gene tree does not necessarily reflect the species tree. • Common ancestor or two gene lineages can predate species split: ancestral polymorphism. • Trees derived from different genes or linkage groups may conflict. Peters, McCracken, et al. (unpubl. data) Hybridization • Gene flow between species or populations also may obscure the species tree: The other big problem: • Recombination complicates phylogenetic inference. Speciation and species concepts: Allopatric model • If populations become subdivided, allele frequencies naturally change over time and and populations diverge. • If or when populations reunite, they may fail to mate or produce inviable offspring allopatric speciation. • Or they might introgress and hybridize; the degree of speciation depends on pre- and post-zygotic barriers: Biological Species Concept Systematics and the Origin of Species Ernst Mayr (1942) Allopatric Model Peripatric Model vicariance Vicariance results in subdivision and through time leads to two reproductively isolated species clades evolving on different trajectories . Migration event followed by peripheral isolation (founding population persists; new daughter species buds off). Types of barriers to gene flow: • Spatial, temporal, and ecological isolation • Post-zygotic barriers • • • Hybrid sterility/inviability • Haldane’s rule: sterility and inviability occurs more often in the heterogametic sex (e.g., because deleterious alleles are exposed on the Y chromosome). • Hybrid breakdown: inviability occurs some generations later. Pre-zygotic barriers • Behavioral incompatibility • Mechanical isolation (genitalia do not fit together) • Gametic isolation (gametes fail to fuse) Reinforcement • Pre-zygotic barriers evolve to reinforce post-zygotic barriers Speciation: Sympatric/Allochronic models • In some cases, speciation may be driven in the absence of allopatry or peripheral isolation. • Speciation and reproductive isolation can correlate with ecological preferences (sympatric) or timing instead of space (allochronic or heteropatric speciation). • Examples of sympatric speciation are though to be driven primarily by adaptation as opposed to vicariance associated with geography. • Several good examples of sympatric speciation documented: Threespine stickleback (Gasterosteus aculeatus) http://fish.dnr.cornell.edu/nyfish/Gasterosteidae/sticklebackpic.html Sticklebacks inhabit lakes and streams in recently deglaciated habitats, evolved from marine ancestors. Concluding remarks: • No sharp division between phylogenetic systematics and population genetics. • Same forces that give rise to micro-evolutionary patterns we observe are responsible for macro-evolutionary patterns that play out over many generations. • Successful integrated analysis requires basic knowledge of population genetics and phylogenetics.