* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download classification of bacteria
Cell culture wikipedia , lookup
Molecular cloning wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microbial metabolism wikipedia , lookup
Point mutation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
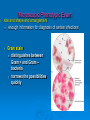
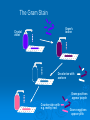
Classification of bacteria DR.THAMINA SAYYED REGISTRAR MICROBIOLOGY KKUH Bacterial cells Classification System 3 Domains 1978 Carl Woese 1. Bacteria • Unicellular prokaryotes with cell wall containing peptidoglycan 2. Archaea • Unicellular prokaryotes with no peptodoglycan in cell wall 3. Eukarya • • • • Protista Fungi Plantae Animalia Comparing Prokaryotic and Eukaryotic Cells Taxonomic Classification Categories arranged in hierarchical order species is basic unit Domain Kingdom Phylum or Division Class Order Family Genus Species Prokaryote Classification Technologies used to characterize and ID prokaryotes microscopic examination culture characteristics biochemical testing nucleic acid analysis combination of the above is most accurate Phenotypic & Genotypic classification Phenotypic Characteristics for Identifying Prokaryotes often does not require sophisticated equipment can easily be done anywhere Microscopic Phenotypic Exam size and shape and arrangement enough information for diagnosis of certain infections Gram stain distinguishes between Gram + and Gram – bacteria narrows the possibilities quickly Microscopic Phenotypic Exam special stain allows for the distinction of microorganisms with unique characteristics • capsule • acid fast staining detects the waxy presence of Mycobacterium tuberculosis Capsule staining Acid fast staining of M. tuberculosis CELL WALL Gram positive cell wall Consists of a thick, homogenous sheath of peptidoglycan 20-80 nm thick tightly bound acidic polysaccharides, including teichoic acid and lipoteichoic acid cell membrane Retain crystal violet and stain purple Gram negative cell wall Consists of an outer membrane containing lipopolysaccharide (LPS) thin shell of peptidoglycan periplasmic space inner membrane Lose crystal violet and stain pink from safranin counterstain 11 Gram Positive Gram Negative 12 The Gram Stain Gram's iodine Crystal violet Decolorise with acetone Gram-positives appear purple Counterstain with e.g. methyl red Gram-negatives 13 appear pink Gram-positive cocci Gram-positive rods Gram-negative cocci Gram-negative rods 15 Metabolic Phenotypic Exam cultural approaches required for positive diagnosis of infection isolation and ID of pathogen accuracy, reliability, and speed methods used include culture characteristics biochemical reactions process Serological Testing Phenotypic Exam serological testing uses ELISA testing fast and easy to use Classification of bacteria Classification of medically significant bacteria I.Thick rigid walled cells A. Free living extracellular 1.Gram positive a.Cocci Staphylococcus - abcess Streptococcus - puemonia, Pharyngitis cellulitis b.Spore forming rods Aerobic Bacillus - Anthrax Anaerobic Clostridium - tetanus,gas gangrene botulism c.Non spore forming rods (GRAM POSTIVE CONTD) 1-Non filamentous Cornybacterium – Diphtheria Listeria - meningitis 2.Filamentous Actinomycetes – Actinomycosis Nocardia - Nocardiosis 2.Gram negative A.Cocci Neisseria -Gonorrhoea, meningitis B.Rods 1.Facultative a. Straight 1.Respiratory org. Haemophillus- meningitis Bordatella-Whooping cough Legionella- Pneumonia 2.Zoonotic Brucella – Brucallosis Francisella –Tularemia Pasteurella –Cellulitis Yersinia - Plague 3.enteric & related (GRAM NEGATIVE CONTD) E.coli - UTI,Diarrhoea Enterobacter – UTI Serratia – Pneumonia Klebsiella – Pneumonia.UTI Salmonella – enterocolitis,typhoid fever Shigella – Enterocolitis Proteus – UTI b. Curved Campylobacter – Entericolitis helicobacter – Gastritis,Peptic ulcer Vibrio - Cholera C.Aerobic D. Anaerobic (Gram negative) Pseudomonas – pneumonia,UTI Bacteroids – peritonitis 3.ACID FAST MYCOBACTERIUM - Tuberculosis & Leprosy B . Non free living obligate intracellular parasites 1.Rickettsia – Rocky mountain spotted fever Typhus, Q fever 2.Chlamydia urethritis, trachoma. Psittacosis Flexible thin walled Spirochaetes - Treponema – Syphilis Borrelia – Lyme disease Leptospira - leptospirosis Wall- less cells Mycoplasma - pneumonia Subtyping & Its applications To distinguishinguish between strains of different species Biotyping Serotyping Antimicrobial susceptibility system Bacteriophage typing Bacteriocin typing Genotypic Characteristics for Identifying Prokaryotes the use of genotypic testing has increased with the availability of technology genotypic testing is particularly useful in the case of organisms that are difficult to identify several techniques include gene probes PCR sequencing rRNA gene probes single stranded DNA that has been labeled with a identifiable tag, such as a fluorescent dye are complementary to target nucleotide sequences • unique in DNA of pathogen Genotypic Characteristics used in Classifying Prokaryotes( non culture methods) PCR: polymerase chain reaction used to detect small amounts of DNA present in a sample (blood, food, soil) the PCR chain reaction is used to amplify the amount of DNA present sequencing ribosomal RNA of particular use for identifying prokaryotes impossible to grow in a culture focus is place on the 16S molecules of the RNA because of it’s size • approximately 1500 nucleotides once the 16S molecule is sequenced, it can then be compared to the sequences of known organisms Genotypic Characteristics used in Classifying Prokaryotes comparison of nucleotide sequences differences in DNA sequence can assist in determination of divergence of evolutionary path for organisms DNA hybridization single strands of DNA anneal 16S ribonucleic acid comparing sequence of ribosomal RNA relatedness to other organisms can be determined using numerical taxonomy determined by the percentage of characteristics two organisms have in common