* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download SVD and PCA COS 323
DNA supercoil wikipedia , lookup
Epigenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
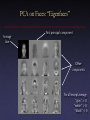
SVD and PCA COS 323 Dimensionality Reduction • Map points in high-dimensional space to lower number of dimensions • Preserve structure: pairwise distances, etc. • Useful for further processing: – Less computation, fewer parameters – Easier to understand, visualize PCA • Principal Components Analysis (PCA): approximating a high-dimensional data set with a lower-dimensional linear subspace * * Second principal component * * First principal component * * * * * * * ** * * Original axes * * ** * * * * * Data points SVD and PCA • Data matrix with points as rows, take SVD – Subtract out mean (“whitening”) • Columns of Vk are principal components • Value of wi gives importance of each component PCA on Faces: “Eigenfaces” Average face First principal component Other components For all except average, “gray” = 0, “white” > 0, “black” < 0 Uses of PCA • Compression: each new image can be approximated by projection onto first few principal components • Recognition: for a new image, project onto first few principal components, match feature vectors PCA for Relighting • Images under different illumination [Matusik & McMillan] PCA for Relighting • Images under different illumination • Most variation captured by first 5 principal components – can re-illuminate by combining only a few images [Matusik & McMillan] PCA for DNA Microarrays • Measure gene activation under different conditions [Troyanskaya] PCA for DNA Microarrays • Measure gene activation under different conditions [Troyanskaya] PCA for DNA Microarrays • PCA shows patterns of correlated activation – Genes with same pattern might have similar function [Wall et al.] PCA for DNA Microarrays • PCA shows patterns of correlated activation – Genes with same pattern might have similar function [Wall et al.] Multidimensional Scaling • In some experiments, can only measure similarity or dissimilarity – e.g., is response to stimuli similar or different? – Frequent in psychophysical experiments, preference surveys, etc. • Want to recover absolute positions in k-dimensional space Multidimensional Scaling • Example: given pairwise distances between cities – Want to recover locations [Pellacini et al.] Euclidean MDS • Formally, let’s say we have n × n matrix D consisting of squared distances dij = (xi – xj)2 • Want to recover n × d matrix X of positions in d-dimensional space 0 ( x1 − x2 ) 2 D= 2 ( x1 − x3 ) ( x1 − x2 ) 2 ( x1 − x3 ) 2 0 ( x2 − x3 ) 2 ( x2 − x3 ) 2 0 ( x1 ) X = ( x2 ) Euclidean MDS • Observe that d ij2 = ( xi − x j ) 2 = xi − 2 xi x j + x j 2 2 • Strategy: convert matrix D of dij2 into matrix B of xixj – “Centered” distance matrix – B = XXT Euclidean MDS • Centering: – Sum of row i of D = sum of column i of D = si = 2 2 2 d = x − 2 x x + x ∑ ij ∑ i i j j j j = nxi2 − 2 xi ∑ x j + ∑ x 2j j j – Sum of all entries in D = s = ∑ si = 2n∑ x − 2 ∑ xi i i i 2 i 2 Euclidean MDS • Choose Σxi = 0 – Solution will have average position at origin – Then, si = nxi2 + ∑ x 2j , s = 2n∑ x 2j j d ij2 − 1n si − 1n s j + j 1 n2 s = −2 xi x j • So, to get B: – – – – compute row (or column) sums compute sum of sums apply above formula to each entry of D Divide by –2 Euclidean MDS • Now have B, want to factor into XXT • If X is n × d, B must have rank d • Take SVD, set all but top d singular values to 0 – – – – Eliminate corresponding columns of U and V Have B3=U3W3V3T B is square and symmetric, so U = V Take X = U3 times square root of W3 Multidimensional Scaling • Result (d = 2): [Pellacini et al.] Multidimensional Scaling • Caveat: actual axes, center not necessarily what you want (can’t recover them!) • This is “classical” or “Euclidean” MDS [Torgerson 52] – Distance matrix assumed to be actual Euclidean distance • More sophisticated versions available – “Non-metric MDS”: not Euclidean distance, sometimes just inequalities – “Weighted MDS”: account for observer bias