* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download O`Brien et al. 1983. The cheetah is depauperate in genetic variation
Dual inheritance theory wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Designer baby wikipedia , lookup
Genetic code wikipedia , lookup
Genetic studies on Bulgarians wikipedia , lookup
Medical genetics wikipedia , lookup
Behavioural genetics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Quantitative trait locus wikipedia , lookup
History of genetic engineering wikipedia , lookup
Koinophilia wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Genetic engineering wikipedia , lookup
Inbreeding avoidance wikipedia , lookup
Genetic testing wikipedia , lookup
Public health genomics wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic drift wikipedia , lookup
Microevolution wikipedia , lookup
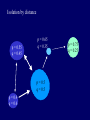
Genetic diversity among populations (and factors that increase or decrease diversity) Populations that are spatially isolated will tend to diverge genetically • genetic drift • natural selection and local adaptation p = .5 q = .5 p = .7 q = .3 p = .55 q = .45 p = .5 q = .5 p = .8 q = .2 Genetic diversity among populations • Increases due to isolation, followed by – genetic drift – inbreeding – selection – local adaptation • Decreases due to gene flow (migration) as migrants move between populations, they homogenize allele frequencies among populations • larger populations diverge slowly through drift – few migrants needed to counteract • small populations diverge rapidly through drift – more migrants needed to counteract Changes in genetic diversity among populations m = proportion of population that migrates Nm = number of migrants randomly exchanged per generation Ne = 1,000, m = 0.01, Nm = 10 Ne = 100, m = 0.01, Nm = 1 approx 1 migrant/generation will maintain same alleles among populations (= qualitative similarity) but 10 migrants per generation may still permit significant differences in allelic frequencies ( = quantitative dissimilarity) Changes in genetic diversity among populations m = proportion of population that migrates Nm = number of migrants randomly exchanged per generation Ne = 1,000, m = 0.01, Nm = 10 Ne = 100, m = 0.01, Nm = 1 “in the absence of natural selection, the amount of genetic divergence among demes is a function of the absolute number of migrants exchanged (Nm), not the proportion of exchange (m)” (Allendorf 1983) Isolation by distance p = 0.55 q = 0.45 p = 0.65 q = 0.35 p=0.5 p = 0.5 q=0.5 q = 0.5 p = 0.4 q = 0.6 p = 0.75 q = 0.25 Isolation by distance http://www.youtube.com/watch?v=31qBrRawDK8 Isolation by distance Mussel ‘lures’ with glochidia Genetic distance between populations Elliptio dilatata Clegg, S.M., S.M. Degnan, J. Kikkawa, et al. 2002. Genetic consequences of sequential founder events by an island-colonizing bird. PNAS 99:8127-8132 silvereye (Zosterops lateralis) Issues with genetic diversity among populations outbreeding depression/hybridization • local adaptation Issues with genetic diversity among populations outbreeding depression/hybridization • local adaptation example: ibex extirpated from Czechoslovakia (Capra ibex ibex) - transplanted from Austria successfully (Capra ibex ibex) - then added bezoars (C. i. aegagrus) and Nubian ibex (C. i. nubiana) - fertile hybrids rutted in early fall instead of winter (as natives did) - kids of hybrids born in February, coldest month of year, entire population went extinct David Hall Issues with genetic diversity among populations outbreeding depression/hybridization • local adaptation • co-adapted gene complexes Loss of fitness due to Inbreeding: accumulation of homozygous recessives loss of superior heterozygotes Outbreeding: reduced fitness of F1 generation - disruption of local adaptation - epistatic interactions reduced fitness of F2 generation - breakup of co-adapted gene complexes More empirical studies on inbreeding than outbreeding…. Reproductive success inbreeding depression inbreeding outbreeding depression random mating inter-breeding Why do we care? Inbreeding: may be last recourse for endangered population Outbreeding: dangerous consequence of saving small popns. “Genetic pollution” controversy Florida panther: declined due to habitat loss, poaching, road kills evidence of inbreeding: low fertility, sperm abnormalities, cowlicks, kinked tails, cardiac defects, undescended testicles, high disease rate Florida panther: Genetic studies indicated low variability: Florida Western US Other felids P 4.9 9.9 8-21 H 1.8 4.3 3-8 DNA H 10.4 29.7 45.9 Found to have hybridized with S. American subspecies; introgressed animals with higher P Florida panther: Outbred with sub-species from Texas - added 8 females in 1995 F1 hybrid kittens do not have cowlinks or kinked tails Texas genes are now 15-29% of total Cheetah (Acinonyx jubatus): O’Brien et al. 1983. The cheetah is depauperate in genetic variation - using protein electrophoresis Species Drosophila Mus Homo sapiens Felis catus Cheetah # popns. N 43 >100 2 87 many >100 1 56 2 55 # loci 24 46 104 55 47 % poly. loci 43.1 20.5 31.7 22 0.0 av. H 0.14 0.088 0.063 0.076 0.0 Cheetah (Acinonyx jubatus): Menotti-Raymond and O’Brien et al. 1993. - using protein electrophoresis, high-resolution PE, and mtDNA Species Drosophila Mus Homo sapiens Felis catus Cheetah # popns. N 43 >100 2 87 many >100 1 56 2 55 # loci 24 46 104 55 47 % poly. loci 43.1 20.5 31.7 22 0.0 av. H 0.14 0.088 0.063 0.076 0.0 Drosophila Mus Homo sapiens Cheetah 1 1 1 1 20 54 72 many 155 11.1 4.1 1.2 3.2 0.04 0.02 0.063 0.013 Felis catus 1 17 46.0 Cheetah 3 76 45.0 34 6 O’Brien et al. 1983. The cheetah is depauperate in genetic variation - assumed to be result of small N, bottleneck, then inbreeding - highly vulnerable to disease outbreaks (50% mortality in one captive population) O’Brien et al. 1983. The cheetah is depauperate in genetic variation - assumed to be result of small N, bottleneck, then inbreeding - highly vulnerable to disease outbreaks (50% mortality in one captive population) Merola, 1994. A reassessment of homozygosity …. - carnivores tend to show low levels of genetic variation (several have lower levels of H and P than cheetah) - measures of fluctuating asymmetry indicate cheetah is not suffering from low homozygosity or genetic stress - sperm deformities – do not affect fertility, may be normal in felids - low litter sizes – in captivity (high in wild) - susceptibility to disease – may be due to captive contact (in wild, cheetahs avoid conspecifics) Concluded that conservation is better directed at habitat