* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download ppt - University of Illinois Urbana
Survey
Document related concepts
Transcript
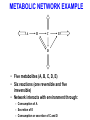
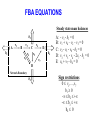
BIOINFORMATICS ON NETWORKS Nick Sahinidis University of Illinois at Urbana-Champaign Chemical and Biomolecular Engineering MOTIVATION • Genomics and proteomics help us understand the structure, properties, and function of single genes and proteins • Genes and proteins function in complex networks • Bioinformatics on biochemical networks aims to understand and rationally manipulate networks of genes and proteins • These networks are very complex – http://www.expasy.org/cgi-bin/show_thumbnails.pl – http://www.expasy.org/cgi-bin/show_thumbnails.pl?2 – http://www.genome.ad.jp/kegg/pathway.html LEARNING OBJECTIVES (two lectures) • Introduction to: – – – – – Metabolic networks Flux balance analysis S-systems theory Gene additions and deletions Pathway reconstruction from data METABOLIC NETWORKS • Definitions – Metabolic network: a system of interacting proteins and small molecules converting raw materials to energy and other useful substances in a living organism – Metabolites: materials consumed or produced in a metabolic network – Enzymes: proteins that catalyze reactions – The sets of metabolites and enzymes of a network are not necessarily disjoint • Key observation – A large proportion of the chemical processes that underlie life are shared across a very wide range of organisms GRAPHICAL REPRESENTATION • Nodes represent metabolites and enzymes • Arcs correspond to reactions and modulation • Dotted or colored lines often reserved to denote modulation • A negative sign associated with an arc is used to denote inhibition METABOLIC NETWORK EXAMPLE A B C E D • Five metabolites (A, B, C, D, E) • Six reactions (one reversible and five irreversible) • Network interacts with environment through: – Consumption of A – Secretion of E – Consumption or secretion of C and D FLUX BALANCE ANALYSIS • Pseudo steady-state hypothesis: metabolic dynamics are much faster compared to those of the environment • Model network through steady-state mass balances for metabolites • For each metabolite, its rate of consumption must equal its rate of production FBA EXAMPLE Internal Fluxes b2 A v1 B v2 b1 v3 v6 C v4 E b4 v5 v7 v1 : A v2 : B v3 : B v4 : D v5 : C v6 : C v7: 2D B C D B D E E D Exchange Fluxes Network Boundary b3 b1: b2: b3: b4: A C D E Exchange fluxes may be positive (system output) or Negative (input to metabolic network) FBA EQUATIONS Steady state mass balances b2 A v1 B v2 b1 v3 v6 C v4 E b4 v5 v7 D A: B: C: D: E: - v1 - b1 = 0 v1 + v4 – v2 – v3 = 0 v2 - v5 - v6 - b2 = 0 v3 + v5 - v4 - 2v7 - b3 = 0 v6 + v7 - b4 = 0 Network Boundary b3 Sign restrictions 0 v1,…,v7 b1 0 - b2 + - b3 + b4 0 MODELING WITH FBA • Problem #1: Interpret metabolic network behavior – Hypothesis: Network is an optimizer – Likely objectives: » Maximize growth » Minimize energy consumption – Leads to a linear program • Problem #2: Manipulate a metabolic network to produce certain desired products through – Control of external fluxes – Structural manipulations in the network GENE ADDITIONS AND DELETIONS • Two-level problem – Upper level: maximize a bioengineering objective through gene knockouts – Lower level: cell is still an optimizer that seeks to optimize its own objective through adjusting internal fluxes • Use binary variable for each gene to decide whether to knock it out or not (or whether to over-express) • Inner linear program can be converted to a set of linear equalities and inequalities via duality theory giving rise to a mixed-integer linear program for the overall problem REFERENCES AND FURTHER READING • B. Palsson, 2000 Hougen Lectures – http://gcrg.ucsd.edu/presentations/hougen/hougen.htm • E. Voit, Computational Analysis of Biochemical Systems, Cambridge University Press, 2000. • N. Friedman, Inferring cellular networks using probabilistic graphical models, Science, 303, 799-805, 2004. • Metabolic Systems Engineering course: – http://archimedes.scs.uiuc.edu/courses/meteng.html












![CLIP-inzerat postdoc [režim kompatibility]](http://s1.studyres.com/store/data/007845286_1-26854e59878f2a32ec3dd4eec6639128-150x150.png)








