* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Tools for functional annotation
Survey
Document related concepts
Transcript
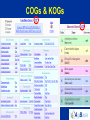
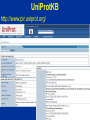
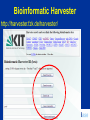
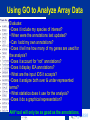
Modeling Functional Genomics Datasets CVM8890-101 Lesson 3 13 June 2007 Fiona McCarthy Lesson 3: Tools for functional annotation. Accessing functional data; computational strategies to obtain more complete functional annotation; the AgBase GO annotation pipeline. Lesson 3 Outline 1. Review: Functional Annotation 2. Tools for functional annotation – Accessing functional data – Computational strategies to obtain more functional data 3. Example: The AgBase GO annotation pipeline 4. Other GO annotation tools Review: Functional Annotation • biologists refer to both the annotation of the genome and functional annotation of gene products: “structural” AND “functional” annotation • Functional annotation is required to make biological sense of high throughput datasets eg. genomics, arrays, proteomics • COGs, KOGs, GO Tools for Functional Annotation • Need to be able to access functional annotation for your dataset – Breadth and depth – Date updated – No annotation vs function unknown • Need to be able to add more annotation • Need to be able to use the annotations to model your data – Depth or detail – Compatibility with other programs (eg pathway analysis) – Comparative data? Tools for Functional Annotation • • • • • • • Clusters of Orthologous Groups (COGs) euKaryotic Orthologous Groups (KOGs) UniProt Knowledgebase (UniProtKB) Bioinformatic Harvester FANTOM Puma Gene Ontology (GO) COGs & KOGs • Accessible at http://www.ncbi.nlm.nih.gov/COG/ • ftp download • Available for many prokaryotes and 7 eukaryotes • Add more annotation using the KOGinator? • Modeling: – Has breadth but not always depth – Good for prokaryote comparative analysis? COGs & KOGs COGs & KOGs http://www.ncbi.nlm.nih.gov/COG/ Automated tools for large numbers of comparisons?? UniProtKB • Accessible at http://www.pir.uniprot.org/ • ftp download & sophisticated search & download capabilities • Available for > 132,000 species • Annotation across both literature (for selected species) and biological databases • Modeling: – Has breadth but not always depth; many proteins not represented in UniProtKB – Those that are represented have a detailed summary of function from a range of sources – Rapid help and feedback from the database help UniProtKB http://www.pir.uniprot.org/ UniProtKB http://www.pir.uniprot.org/ UniProtKB http://www.pir.uniprot.org/ Bioinformatic Harvester • Accessible at http://harvester.fzk.de/harvester/ • no download • Available for 6 model species • Integrates data from multiple sources • Modeling: – Has breadth and depth; not useful for large datasets – Updates? Bioinformatic Harvester http://harvester.fzk.de/harvester/ FANTOM http://www.gsc.riken.go.jp/e/FANTOM/ Mouse only PUMA http://compbio.mcs.anl.gov/puma2/ Gene Ontology • Accessible at http://www.geneontology.org/ • updated downloads for 34 species + downloads for UniProtKB species (>130,000) • UniProtKB species annotation: some depth, less breadth • GO data mapped from other databases • Modeling: – Many tools available for modeling using the GO – Can use computational or manual curation to add annotations Gene Ontology http://www.geneontology.org/ Accessing GO Data EBI-GOA Project http://www.ebi.ac.uk/GOA/ The AgBase GO Annotation Pipeline • Accessible at http://www.agbase.msstate.edu/ • Access available annotations for agriculturally important species • Provide your own GO annotations • Model GO for your dataset Coming soon; GOModeler quantitative hypothesis driven modeling using GO Other GO Annotation Tools http://www.geneontology.org/GO.tools.shtml Other GO Annotation Tools Evaluate: • Can I run it from my computer? • Does it include my species of interest? • When was it last updated? • Does it display evidence codes? • Does it display IEA annotations? • What are the inputs it accepts? • Does it do batch searches? Using GO to Analyze Array Data Using GO to Analyze Array Data Evaluate: • Does it include my species of interest? • When were the annotations last updated? • Can I add my own annotations? • Does it tell me how many of my genes are used for the analysis? • Does it account for “not” annotations? • Does it display IEA annotations? • What are the input IDS it accepts? • Does it analyze both over & under-represented terms? • What statistics does it use for the analysis? • Does it do a graphical representation? ANY tool will only be as good as the annotations.