* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download rnai_presentation
Survey
Document related concepts
Transcript
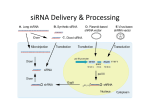
Kyle Breakey, Kylie Broadbent, Christine Mineart, Matt Perez http://www.life.uiuc.edu/shapiro/RNAipathway.jpg Limitations Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans B. Sönnichsen1, L. B. Koski1,6, A. Walsh1, P. Marschall1,6, B. Neumann1,6, M. Brehm1, A.M. Alleaume1,6, J. Artelt1,6, P. Bettencourt1,6, E. Cassin2,6, M. Hewitson1, C. Holz1, M. Khan1, S. Lazik1, C. Martin1, B. Nitzsche1,6, M. Ruer2, J. Stamford2, M. Winzi1, R. Heinkel1,6, M. Röder1,6, J. Finell1,6, H. Häntsch1, S. J. M. Jones3, M. Jones4,6, F. Piano5, K. C. Gunsalus5, K. Oegema2,6, P. Gönczy2,6, A. Coulson4,6, A. A. Hyman2 and C. J. Echeverri1 2005 2000 http://www.newsdesk.umd.edu/scitech/images/worm-briggsae_WT_male.gif Limitations • Endpoint Analysis: (older) – Phenotypes scored when researcher made observations – Phenotypes were missed-such as delays in cell division – Can’t differentiate between secondary effects and the direct knockdown of genes • Movies: (less old) – DIC relies on manual phenotyping • Biased and not quantitative • Expensive and time consuming • Live-Cell Assay: (2006) – Image captured every 30 min. and made into a video clip – Computerized analysis of phenotypes using fluorescence imaging HeLa? • Henrietta Lacks Died of cervical cancer on the 4th October, 1951 at Johns Hopkins University Hospital. • Biopsy Sample sent for analysis and deemed malignant – Then revealed to be the 1st immortal human cell line. • Immortal Capable of dividing outside of the body an unlimited number of times as long as fundamental survival conditions are sustained. • Ethical? Went to the supreme court who ruled that discarded tissue and cells are no longer the property of the individual and are free to be commercialized… http://www.hno.harvard.edu/gazette/2001/07.19/photos/4-Gilbert-250.jpg HeLa? • Instrumental in creating the Polio vaccine HeLa Karyotype • May be the key to curing cancer? • Karyotype shows 84 chromosomes – Thought to have fused with HPV genome through horizontal gene transfer http://cancerres.aacrjournals.org/cgi/content/full/59/1/141#SEC6 siRNA Cell Array (See references #8-9) http://www.nature.com/ng/journal/v37/n6s/full/ng1560.html • siRNA-gelatin transfection solution was prepared in 384 plates. – Transfection=the introduction of foreign material into eukaryotic cells • HeLa cells seeded on top of the array • Spotted siRNA are taken up from the solid phase by the cells morphology Reporter expression and immunostaining Viability/cell count -49 target genes known to be related to chromosome segregation or nuclear structure -synthesized one siRNA for each gene and assayed on HeLa cells dsRNAi sequences used -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells - printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -43/49 gene knockdown efficiencies measured by qRT-PCR -all siRNAs showed partial knockdown dsRNA primers used to quantify expression levels of targeted gene products. Supplementary Figure 1, Neumann et al. relative remaining mRNA (%) siRNA knock-down efficiency. mRNA knock-down efficiency of 43 siRNAs targeting 42 of the 49 endogenous genes with predicted cell cycle function as well as the peripheral Golgi coatamer protein COPB as a known suppressible gene. For the remaining six siRNAs qRT-PCR did not yield products due to difficulties in primer design. mRNA expression levels were determined 48 hours after transfection and are shown as average and standard deviations of three independent experiments. -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -43/49 gene knockdown efficiencies measured by qRT-PCR -all siRNAs showed partial knockdown -scored phenotypes by fluorescence microscopy Figure 1. Workflow of highthroughput RNAi screening by time-lapse imaging. Flowchart of the steps, including time lines of each process based on one live-cell microarray containing 384 siRNAs. -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -43/49 gene knockdown efficiencies measured by qRT-PCR -all siRNAs showed partial knockdown -scored phenotypes by fluorescence microscopy -TPX2 and COPB RNAi knockouts -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -43/49 gene knockdown efficiencies measured by qRT-PCR -all siRNAs showed partial knockdown -scored phenotypes by fluorescence microscopy -TPX2 and COPB RNAi knockouts -can store microarrays for 7 months w/o loss of transfection -49 target genes known to be related to chromosome structure -synthesized one siRNA for each and assayed on HeLa cells -printed automatically prepared ‘transfection cocktails’ for 50 siRNAs into imaging chambers -siRNA taken up by sample cells -43/49 gene knockdown efficiencies measured by qRT-PCR -all siRNAs showed partial knockdown -scored phenotypes by fluorescence microscopy -TPX2 and COPB RNAi knockouts -can store microarrays for 7 months w/o loss of transfection - Scrambled siRNA samples did not affect cell growth -http://www.nature.com/nmeth/journal/v3/n5/suppinfo/nmeth876_S1.html Automated Phenotype Analysis • To begin this process, the boundaries of the fluorescent chromosomes were defined by segmentation using “optimized local adaptive thresholding” – Histones were tagged with GFP – An algorithm is developed to segment the image to be studied – Segmentation is based on the gray levels in the pixels of the image • This identified interphase nuclei and mitotic chromosome sets with over 99% accuracy (compared to manual analysis) • Second, the texture and shape properties of each chromosome set were computed. • Extracted numerical features from each segmented chromosome and defined values: – Shape – Texture • From these properties of texture and shape, biological classes were defined • The classes defined: – Interphase – Mitosis – Apoptosis • Several different cell-death phenotypes – Shape • Nuclei of abnormal shape • Automatic classification into these classes was done by machine-learning – multiclass SVMs • SVM - support vector machine – Vectors are used to “teach” the classifier to identify and classify images into classes – In this example, vectors that defined shape and texture features from the chromosome sets were used to teach the SVM to identify different biological classes • The SVM classifier was trained on the 45 bestranked features of 1,000 manually labeled cells – Classifier achieved 97% accuracy on all samples • There were 89 frames per movie for each siRNA, for every single frame of the time-lapse movie, of each spotting position, a class was assigned to the cells • This gave plots of the change in phenotype over time • They used automated phenotyping for the analysis of these images as it allowed for a “quantitative, unbiased, and reproducible analysis of phenotypes” – A project like this generates lots of data, to efficiently analyze it all a method like this had to be developed • Segmentation and classification analyses were done for 2 normal cell divisions on a scrambled siRNA spot and the software correctly assigned the interphase and mitotic chromosomes. • Apoptotic cells were visualized by suppressing nuclear envelope proteins – SYNE2 and PLK1 • Knock-down of an inner centromere protein INCENP resulted in segregation defects that were correctly assigned to the shape class • An entire movie was analyzed for classification – For siSYNE2, siPLK1, siINCENP – Plots of “mitotic”, “shape”, and “apoptotic” analyses were generated • Each analysis was done by taking the number of the cells in the area of the movie that corresponded to a class and that was compared to the total number of cells in that same area. • The percentage of cells in a class were compared to the data generated for the scrambled siRNA cells. • If any of the three classes were significantly different from the scrambled siRNA cells (control), the phenotype appearance and phenotype penetrance were defined. • By observing the effects of a specific gene knockdown over time, more detailed information regarding the phenotype of the knock down could be gathered – Phenotype penetrance • The maximum difference from the control – Phenotype appearance • the earliest time at which the difference from the control cells Automated Phenotype Analysis • The software developed to observe the phenotype of the cells performed: • 1. Found location of chromosomes in each cell for each frame • 2. Classified the chromosomes by morphology • 3. Detected cell-cycle phenotypes based on classification results for an entire movie • This method developed a way to analyze a genome-wide RNAi study efficiently and accurately Quality Analysis • Compared automatic phenotyping (Computer) to manual phenotyping (Human) • For mitotic phenotypes: 86% agreement (43/49) – 14% false postive (7/49) • Apoptotic phenotypes: 88% agreement (44/49) – 10% false postive (5/49) Phenotypes Defined • Mitotic – Abnormalities in cell division • Shape – Unusual shape of nucleus • Apoptosis - DEATH Classification of Genes • Phenotype displayed -Mitotic - Shape - Apoptotic • Penetrance - Used the mean of the maximum values Classification of Genes (cont.) • Order of appearance of phenotypes – Not possible with endpoint assays – Increases ability to categorize • Indicates stability of targeted protein and efficiency of siRNA Multiparameter Clustering • Grouped genes by phenoprint – Phenotype observed over time This is cool but… • Machine learning has limitations – Only recognize phenotypes it was programmed to recognize – Explains 14% false postive rate • Lots of programming to recognize phenotype of all the different cell types Important to Remember • The point of this paper was to design a high throughput method of detecting phenotypes using RNAi – Not to discover the specific phenotypes of the genes used in the experiment 49 RNAi constructs were used to assay mitotic assembly Time-lapse imaging resolved phenotypes developed a method for high-throughput RNAi data analysis clear advantages for time-lapse imaging over end-point analysis detects mitotic delays, rather than just mitotic arrest The End (Apoptosis) Questions?