* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Inducing Apoptosis of Glioblastoma Multiforme Cells Using a Bcl
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
RNA interference wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
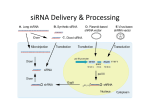
Inducing Apoptosis of Glioblastoma Multiforme Cells Using a Bcl-2 Specific siRNA Sequence Valerie P. Capozziello* and Jeffrey P. Thompson York College of Pennsylvania, Department of Biological Sciences Glioblastoma multiforme (GBM) is the most common brain cancer of middle aged Americans. Unfortunately, survival rates are typically less than 1 year from time of diagnosis. Following surgery, GBM patients typically endure chemical and radiotherapies to slow tumor regrowth. Research suggests that under these treatments GBM cells regress by entering apoptosis, or programmed cell death. We investigated the use of small interfering RNAs (siRNAs) to induce apoptosis in human GBM cells in vitro. Current research suggests that siRNAs eliminate a particular target protein from a cell by degrading the target’s mRNA cellular pool. Bcl-2 is an anti-apoptotic protein in cells. Using Bcl-2 targeted siRNAs, we attempted to eliminate Bcl-2 from U87mg (human GBM) cells, cultured in vitro, inducing them into apoptosis. Preliminary data suggests that U87mg cell number is reduced (cell death enhanced) when treated with Bcl-2 siRNA compared to U87mg control cells, treated with a scrambled siRNA sequence. This research aims to investigate the use of siRNAs to downregulate the expression of the Bcl-2 gene and thereby induce apoptosis of U87MG Human GBM cells (Figure 1). Materials and Methods The suppression of genes by siRNA is a pathway that begins with double-stranded RNA (dsRNA) and is a two-step process (Figure 2). •RNA interference (RNAi) machinery detects dsRNA and breaks it into approximately 21-23 nucleotide long segments. • These segments are bound in an RNA-protein complex that mediates the detection and destruction of mRNAs containing the same sequence as the dsRNA (Pierce 2002). Figure 2. Introduction Glioblastoma multiforme (GBM) is the most common brain cancer among middle aged Americans and has a mean survival rate of approximately one-year (Julien et al. 2000). Cancer is the unregulated growth and proliferation of cells. Cell growth is a process normally controlled by apoptosis. Apoptosis is programmed cell death brought on by a variety of stimuli. This process involves decreased cell volume, modification of the cytoskeleton and degradation of DNA. After this occurs the cell releases tiny membrane-bound apoptotic bodies containing intact organelles that are quickly phagocitized. A new set of oncogenes is being defined that affect this apoptotic pathway (Julien et al. 2000). One of these is the Bcl-2 gene. Bcl-2 does not necessarily affect proliferation but it does inhibit apoptosis (Reed 1994). Bcl-2 has been found to be overexpressed in GBM (Julien et al. 2000). Not only does Bcl-2 affect apoptosis but also increased production of the gene has been found to increase resistance to chemotherapies and radiation treatment, therapies aimed at inducing apoptosis (Reed 1994). Figure 1. Figure 1. U87MG Human GBM cells. Experiments Aseptically transferred 4x105 dilution of U87MG cells into a 96-well cell culture plate Transfected cells with siRNA and Transmessenger Transfection reagent complex Optimization reaction was performed to access the correct ratio of siRNA to TransMessenger to allow the most transfection Target sequence experimental siRNA and scrambled sequence control siRNA were used along with cells untreated with siRNA Efficiency of transfection was measured by reading the fluorescence of the cells after incubation with siRNA complexes Cells were transfected with siRNA and Transmessenger Transfection Reagent complexes at optimal ratio with a larger sample size Transfection was accessed in target sequence and scrambled sequence cells 2500 Fluorescence (485nm/535nm, 0.1s) Objective 2250 2000 1750 1500 1250 1000 750 500 250 0 1:3 1:6 1:12 Target Sequence Figure 3. Mean fluorescence of U87MG cells after transfection. Three ratios of siRNA to Transmessenger Transfection reagent were studied for both the target and scrambled sequence. The 1:12 ratio had the highest transfection rate in both cases. Figure 4. 1.0×10 -04 9.0×10 -05 8.0×10 -05 7.0×10 -05 6.0×10 -05 5.0×10 -05 4.0×10 -05 3.0×10 -05 2.0×10 -05 1.0×10 -05 0.0×10 -00 Cell viability was accessed by incubating cells with cell proliferation reagent PMS-MTS Change in absorbance of the reagent quantified amount of cell viability Standardized cell viability to amount of transfection to calculate cell death using the following calculation Absorbance of control– Absorbance of experimental Fluorescent units Figure 2. siRNA mechanism of gene suppression. •Sequence specific siRNAs can now be readily ordered to match a particular part of a gene sequence. •Instructions for picking a fragment were obtained from xeragon.com, the same company from which the siRNAs were ordered. •The target sequence was verified to be found in the Bcl-2 gene and not in any other gene sequence. •The gene fragment of the Bcl-2 gene 5’-AACATCGGCCCTGTGGATGACTG-3’ was chosen according to those guidelines. •In addition, siRNA was tagged with flourescein on the 3’-end, during synthesis by the company, for identification and quantification of transfection. Results 1. Optimal concentration of siRNA to Transmessenger Transfection reagent was 1:12 for both the target sequence and the scrambled sequence (Figure 3). 2. Transfection efficiency was greater in the scrambled versus target sequence. 3. Target sequence caused approximately three times the amount of cell death compared to scrambled sequence (Figure 4). 4. Standardization was required because of differing amounts of transfection between target and scrambled sequence. 1:3 1:6 1:12 Scrambled Sequence Ratio of siRNA duplex to Transmessenger Reagent Absorbance/Fluorescent Unit Abstract Figure 3. Target Sequence Scrambled Sequence siRNA Figure 4. Calculated amount of cell death in U87MG cells after transfection and cell viability assay. Discussion This data suggests that the siRNA mechanism is a possible method to induce cell death in human GBM cells. Efficiency of transfection is affected by ratio of siRNA to Transmessenger Transfection reagent and this must be optimized. Amount of transfection was also affected by differing uptake by cells even though they were presented with the complexes in the same manner. This is why standardization of cell death was required. Further study is needed to determine whether the observed death is due to apoptosis. Future Directions 1. Perform Western blot to determine if there is a decrease in the Bcl-2 protein level. 2. Determine if death is through apoptosis by looking for apoptotic markers. Literature Cited http://www.xeragon.com/siRNA_review.html Accessed 9/9/2002 http://www.xeragon.com/siRNA_support.html Accessed 9/9/2002 Julien, T., Frankel, B., Longo, S., Kyle, M., Gibson, S., Shillitoe, E., and Ryken, T. 2000. Antisense-mediated inhibition of the Bcl-2 gene induces apoptosis in the human malignant glioma. Surgical Neurology. 53:360-9. Pierce, J.L. 2002. RNAi turns the message off.The Scientist. 16:44-6. Reed, J.C. 1994. Bcl-2 and the regulation of programmed cell death. The Journal of Cell Biology. 124:1-6. Acknowledgments The Pennsylvania Academy of Science for funding