* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Design of Genetical Genomics Studies Which Use Two
Human genetic variation wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Behavioural genetics wikipedia , lookup
Population genetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Public health genomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
Heritability of IQ wikipedia , lookup
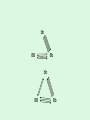
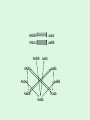
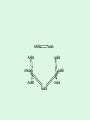
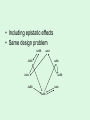
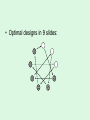
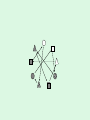
Statistical Analysis and Design of Experiments for Large Data Sets Steven Gilmour School of Mathematical Sciences Centre for Statistics Introduction • I will discuss microarrays, but there are many other possible biological applications • Microarray experiments provide a measure of gene activity • Used to compare expression levels of “treatment” groups • Single channel (e.g. Afymetrix) arrays, or two-colour platforms False Discovery Rate • Hypothesis test procedures for a single response variable are unsuitable for screening for thousands of genes • Testing at 5% level of significance would imply wrongly rejected very large numbers of null hypotheses (declaring inactive genes to be active) • Traditional corrections, such as familywise error rate are too conservative • False discovery rate (FDR) ensures that a suitably small proportion of genes declared active are truly inactive. Sample size calculations • Many methods have been suggested for determining an appropriate number of slides • Assume fixed, unstructured, treatments • Microarrays used recently in genetical genomics studies to understand genetic mechanisms governing variation in complex traits • Treatments now have structure, e.g. family structure, multiloci genotypic groups • We have worked out better sample size methods for such treatments Design for Two-Colour Arrays • Slides are blocks of size two, so incomplete blocks are usually needed • Two colours imply a row-column structure • Designs suggested by several authors • Examples for 4 and 9 treatments Structured Treatment Effects • Three possible genotypes, e.g. F2 populations and codominant markers • Modelled by additive-dominance model • Single locus, genotypes bb, Bb, BB • Plot variance vs. proportion of each homozygous group (r) • Optimal treatment design and blocking for 10 slides: (a) additive effect; (b) dominance effect; (c) both bb BB bb BB Bb bb BB Bb • For multiple loci, factorial structures are used • Two-locus experiment in 10 slides • Optimal treatment design and blocking follow AABB aabb AAbb aaBB AABB aabb AABb aaBb AAbb aaBB AaBB Aabb AaBb AABB aabb AABb aaBb AAbb aaBB AaBB Aabb AaBb • Including epistatic effects • Same design problem AABB aabb AABb aaBb AAbb aaBB AaBB Aabb AaBb Random Treatment Effects • Aim to get good estimates of genetic variances and heritabilities • Designs to find BLUPs of breeding values, given a known pedigree • Two simple pedigree structures: Progeny 1 2 3 4 5 6 7 8 9 Dam 1 2 3 4 5 6 7 8 9 Sire 1 1 1 2 2 2 3 3 3 Dam 1 2 3 1 2 3 1 2 3 Sire 1 1 1 2 2 2 3 3 3 • Optimal designs in 9 slides: Discussion • Consideration of different experimental objectives should lead to different types of design being used • Often a search algorithm is needed to find an optimal design – we have written an R function • There are still many open questions