* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Beili B24 25 Insulin
Signal transduction wikipedia , lookup
Development of analogs of thalidomide wikipedia , lookup
Metalloprotein wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Genetic code wikipedia , lookup
Drug design wikipedia , lookup
Proteolysis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
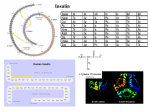
Site Specific Introduction of Unnatural Amino Acids at Sites Critical to Insulin Receptor Recognition & Biological Activity B.Quan, D.Smiley, V.Gelfanov, and R.DiMarchi Department of Chemistry Indiana University, Bloomington Indiana 47405 U.S.A Abstract Experimental Design 60 CH2 3.55 50 49 Insulin 43 LEU LEU 42 GLU VAL LYS 41 GLN ARG A-CHAIN 40 GLY GLY 39 VAL 2 ILE 38 GLN 3 VAL LEU GLU 4 S 5 ASP 36 S GLN CONNECTING PEPTIDE CYS CYS 6 ALA SER 7 8 1 PHE VAL 9 ILE 10 S CYS SER ASN LEU TYR GLN CYS LEU GLU ASN TYR 20 0.51 11 12 13 14 15 16 17 18 19 20 GLN 4 S LEU PRO 6 7 THR S GLY TYR SER 8 HIS LEU 9 10 11 VAL 12 ARG GLU ALA GLU LEU TYR LEU VAL CYS GLY 13 14 15 16 17 18 19 20 21 22 GLY 40 0 CH2 1E-3 0.01 0.1 1 10 16.58 100 0.49 1E-3 0.01 0.1 CF3 [Peptide], nM 1 10 100 1000 [Peptide], nM Fig 3. Binding Affinity of Insulin Analogs with Comparable Amino Acid Substitution at B24 & B25 B24(2-Me) B24(3-Me) B24(4-Me) 200 B24 IC50 nM CH2 18.25 CH2 22.54 CH3 N 60 CH2 40 6.78 CH2 0.63 S Insulin CH3 20 35 CH2 34 B24(4-pyridyl) B24(4-thienyl) 180 80 160 140 120 5.25 PHE PHE 24 1E-3 32 0.01 0.1 1 10 100 80 Results & Conclusion 60 40 Insulin 0 -20 1E-3 CH3 1000 0.01 [Peptide], nM 30 30 0.1 1 10 100 [Peptide], nM 29 28 27 26 Fig 4. Binding Affinity of Insulin Analogs Substituted at Position B24 25 23 Fig 1. Primary Structure of Human Proinsulin B25 IC50 nM B25 IC50 nM 140 >1000 Insulin B25 Phe(4-CF3) Theoretical MW= 5875 0.46 Χ 15cm Vydac C18 1ml/min, 45C, 214nm, Linear gradient of CH3CN in 0.1% TFA(aq) Ubiquitin (M+H)+ average Ubiquitin (M+2H) 2+average CH2 CH2 13.15 B25(4-Me) B25(4-Br) B25(4-CH2NH2) hPhe25 B25(4-COOH) 120 100 CH2 0.67 CH3 80 CH2 60 40 CH2 Insulin 1000 Nineteen different amino acids (17 non-natural) were substituted for the native phenylalanine residues at positions B24 and B25 in insulin. The analogs were successfully prepared by SPPS from individual A and B chains, in total yields that varied between 1 and 10%. The relative binding affinity of this group of insulin analogs for the insulin receptor was determined to differ by more than thousand-fold from the most potent to the least potent peptide studied. Consistent with previous observations predominantly employing native amino acids, we observed that insulin activity was highly dependent upon aromatic character at these two residues (1). Position B24 was extremely restrictive to structural modification while B25 was extremely permissive. In Figure 3 we report the extreme difference between the same substitutions at B24 and B25. Each of the B25 insulin analogs were of high affinity while only one of the B24 analogs did not exhibit a sizable reduction in binding affinity. The highly homologous B24 Phe(4-F) analog is a rare example of an l-amino acid with comparable potency to the native hormone, and represents an opportunity for structural study in the receptor recognition site with isotopically enriched amino acid. Further study at B24 (Figure 4) illustrates that even a single methyl group added to the phenyl ring is deactivating and most notably when ortho to the ring. Phenylalanine heterocycle-based mimetics were studied. An appreciable difference was observed among two commonly used derivatives. The 5-membered thiophene ring provided nearly native insulin affinity but the more alkaline 6-membered pyridine ring was sizably reduced in potency. Additional study at position B25 (Figure 5) demonstrated that movement of the phenyl ring closer or further from the peptide backbone had a significant deactivating impact upon binding affinity (2). Additional modifications that selectively introduced negative charge (4-COOH) and size (4-Br) relative to the native Phe were largely without effect. Although, the alkaline amino-methyl substitution (4-CH2NH2) reduced binding affinity to a significant degree. This observation is consistent with the more subtle reduction noted for the less alkaline 4-NH2 modification (Figure 3). 3.15 20 CH2NH2 0 0.01 0.1 1 10 100 1000 References CH2 0.55 [Peptide], nM Fig 2. Purified Insulin Analog Chromatographic & Mass Spectral Analysis 0.83 COOH 1E-3 Receptor Binding : The binding affinity of each insulin analog for the insulin receptor was measured in a competition binding assay utilizing scintillation proximity assay technology. Serial 3-fold dilutions of the peptides were made in scintillation proximity assay buffer (0.05 M Tris-HCl, pH 7.5, 0.15 M NaCl, 0.1% w/v bovine serum albumin) in a 96 well plate (Corning Inc., Acton, MA) with 0.05 nM (3-[125I]-iodotyrosyl) TyrA14 Insulin. An aliquot of 1-6 micrograms of plasma membrane fragments prepared from cells over-expressing the human insulin receptor and 1 mg/well polyethyleneiminetreated wheat germ agglutinin type A scintillation proximity assay beads (Amersham Biosciences, Piscataway, NJ) were added to each well. The plate was incubated for twelve hours at room temperature and measurements made with MicroBeta1450 liquid scintillation counter (Perkin-Elmer, Wellesley, MA). Non-specifically bound (NSB) radioactivity was measured in the wells with fourfold greater concentration of “cold” native ligand than the highest concentration in test samples and total bound radioactivity was detected in the wells with no competitor. Percent specific binding was calculated as following: % Specific Binding = Bound-NSB / Total bound- NSB x 100. IC50 values were determined by using Origin software (OriginLab, Northampton, MA). 100 20 0 31 THR LYS CYS IGF-1 20 0.91 0 ARG HIS 5 60 Insulin Synthesis of B-chain analogs : B chain analogs with unnatural amino acids at B24 and B25 were successfully synthesized by solid phase methodology using Fmoc chemistry. Peptides were cleaved from the support in strong anhydrous acid and purified under acidic conditions by preparative high performance reverse-phase chromatography in the free sulfhydryl form. Each peptide was characterized by MALDI-MS and HPLC analysis to be at least 90% pure prior to chain combination. Chain Combination Reaction : Each B-chain analog was mixed with a molar equivalent of native A-chain S-sulfonate (gift from Eli Lilly) in a 0.05 M glycine buffer, pH 10.5. A molar equivalent of DTT relative to each S-sulfonate was used to facilitate disulfide interchange. The reaction was conducted at 4ºC, overnight. The product was purified by preparative high performance reverse phase chromatography in a slightly alkaline NH4HCO3 buffer with CH3CN elution. The insulin analogs were obtained in step yield that varied from 5-30%, and characterized by MALDI-MS and HPLC analysis to be greater than 95% pure. 33 21 ARG S 80 F GLU ASN 3 GLU THR B-CHAIN 2 37 NH2 CH2 48 44 1.03 IGF-1 %Specific Binding 1 55 100 0.72 6.79 100 GLN 64 65 56 51 47 LEU SER GLY 59 PRO GLN 46 LEU ALA GLY ALA 60 PRO 45 LEU GLY 61 GLU GLY GLY GLY SER 62 63 57 52 0.67 OH %Specific Binding 58 53 0.67 80 120 54 CH2 CH2 40 B25(4-CF3) B25(4-F) B25(4-NH2) B25 (4-OH) 120 %Specific Binding 100 B25 IC50 nM %Specific Binding Our work has focused on two amino acids in the C-terminus of the Bchain that are central to insulin activity, specifically positions B24 and 25. The selective introduction of a series of Phe-related derivatives at each position has yielded an insightful perspective on insulin receptor recognition. Position 24 has proven highly restrictive to changes that can be introduced and the results emphasize the importance of backbone conformation to full potency. In comparison, position B25 is quite accommodating to sizable changes in side-chain structure but surprisingly highly demanding in that the phenyl ring must be positioned at the beta-carbon. These collective observations establish a foundation for application of unnatural amino acids as a route to insulin pharmacology that may not be obtainable with natural amino acids alone. B24 IC50 nM B24(4-F) B24(4-NH2) B24 (4-OH) B24(4-CF3) 120 %Specific Binding Insulin constitutes a hormone of central importance in physiology and a vital element in glucose management. Its use in diabetes care has been of seminal significance for nearly a century. The advent of rDNA biosynthesis provided human insulin in virtually unlimited quantity. More importantly, it provided a mechanism by which improved biosynthetic insulin analogs could be synthesized, evaluated and registered as new medicines. The advent of chemical biotechnology (biosynthesis with unnatural amino acids) provides a new venue for optimizing insulin pharmacology through the use of synthetic chemistries that otherwise would be prohibitively expense for commercialization. 1. Mirmira, R.G., Nakagawa S.H., and Tager H.S. (1991) J. Biol. Chem. 266, 1428-1436 2. Nakagawa, S. H. And Tager, H.S. (1986) J. Biol. Chem. 261, 7332-7341 Br Fig 5. Binding Affinity of Insulin Analogs Substituted at Position B25 Acknowledgements Beili Quan is financially supported by an Indiana University Gill Center Fellowship.