* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Multi-state modeling of biomolecules wikipedia , lookup
Protein moonlighting wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
List of types of proteins wikipedia , lookup
Biochemistry wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Drug discovery wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Protein domain wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
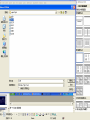
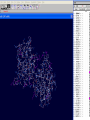
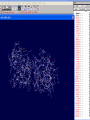
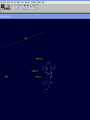
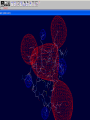
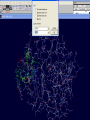
3D structure -Swiss Pdb Viewer David Shiuan Department of Life Science, Institute of Biotechnology and Interdisciplinary Program of Bioinformatics National Dong Hwa University Swiss-PdbViewer a tool for viewing and manipulating protein structures and models. tightly linked to Swiss-Model, an automated homology modeling server. Swiss-PdbViewer provides a user friendly interface allowing to analyze several proteins at the same time. The proteins can be superimposed in order to deduce structural alignments and compare their active sites or any other relevant parts. Amino acid mutations, H-bonds, angles and distances between atoms are easy to obtain thanks to the intuitive graphic and menu interface. Welcome to the SWISS-MODEL Protein Modelling Server Dear David Shiuan, Title of your Request: lipase3D Swiss-Model '(SWISS-MODEL Version 36.0003)' started on 16-Dec-2005 Process identification is AAAa03uOW The modelling procedure is now in progress, and its results should be sent to you shortly. In case of difficulties, please contact: Torsten Schwede Nicolas Guex <[email protected]> News from SWISS-MODEL: - New Server Hardware - DeepView - SwissPDB-Viewer - ExPDB template database Good luck with your projects. The SWISS-MODEL Team Dr. Torsten Schwede Dr. Nicolas Guex Dr. Manuel C. Peitsch [email protected] AlignMaster output ============== Length of target sequence: 288 residues Searching sequences of known 3D structures Found 1f3rB.pdb with P(N)=6e-76 Found 1dzbB.pdb with P(N)=2e-70 Found 1dzbA.pdb with P(N)=2e-70 Found 1nqbC.pdb with P(N)=1e-66 Found 1nqbA.pdb with P(N)=1e-66 Found 1qokA.pdb with P(N)=2e-65 Found 1lmkG.pdb with P(N)=5e-64 Found 1lmkC.pdb with P(N)=5e-64 Found 1lmkE.pdb with P(N)=5e-64 Found 1lmkA.pdb with P(N)=5e-64 Found 1deeC.pdb with P(N)=8e-55 Please find enclosed your results for AAAa03uOW as an attachment. Title : lipase3D Jobid:AAAa03uOW Batch.1 SWISS-MODEL project files are best viewed using the program - DeepView, SwissPdbViewer (http://www.expasy.org/spdbv/). The program allows you to display and manipulate several structures at the same time, e.g. the model and all the superposed templates. How to visualize and analyze the SwissModel generated structures The result file can be visualized by Swiss-pdb Viewer or RasWin (RasMol). Use the Control Panel to remove all of the templates. Select known 3D structures of lipase (for example). Compare the target sequence with the known lipase structure by magic fit of Swiss-pdb Viewer. Use center the molecule on one atom tool bar to focus on the region of binding site. Compare the above two structures. Hermicola (Thermomyces) lanuginaosa lipase (1DU4) Swiss Model – Target (Ab) with 3 templates Target (Ab) model Target and lipase iDU4 Magic Fit – target and lipase 1DU4