* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download (a) (b)
Survey
Document related concepts
Transcript
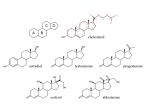
Chapter 2: Chemistry of Life (Chemistry Comes Alive) Niels Bohr – originator of the model of the atom we utilize in BIOL 232 & BIOL 233.. Selman Waksman He investigated how soil microbes defended themselves against invaders which lead to the He and coworkers isolation of twenty-two different defensive compounds produced by soil microbes. These discoveries lead to the discovery streptomycin, the first antibiotic effective against tuberculosis. Waksman received the 1952 Nobel Prize in physiology or medicine. Chemistry still plays a significant role in the cutting-edge research into physiology today. Researchers with deep understanding of chemistry are needed in medicine, physiology, and related fields. Figure 2.1 Two models of the structure of an atom. Nucleus Nucleus Helium atom Helium atom 2 protons (p+) 2 neutrons (n0) 2 electrons (e–) 2 protons (p+) 2 neutrons (n0) 2 electrons (e–) (a) Planetary model Proton Neutron (b) Orbital model Electron Electron cloud Figure 2.2 Atomic structure of the three smallest atoms. Proton Neutron Electron Hydrogen (H) (1p+; 0n0; 1e–) Helium (He) (2p+; 2n0; 2e–) Lithium (Li) (3p+; 4n0; 3e–) Figure 2.3 Isotopes of hydrogen. Proton Neutron Electron Hydrogen (1H) (1p+; 0n0; 1e–) Deuterium (2H) (1p+; 1n0; 1e–) Tritium (3H) (1p+; 2n0; 1e–) Figure 2.5 Chemically inert and reactive elements. (a) Chemically inert elements Outermost energy level (valence shell) complete 8e 2e Helium (He) (2p+; 2n0; 2e–) (b) 2e Neon (Ne) (10p+; 10n0; 10e–) Chemically reactive elements Outermost energy level (valence shell) incomplete 4e 1e Hydrogen (H) (1p+; 0n0; 1e–) 2e Carbon (C) (6p+; 6n0; 6e–) 1e 6e 2e Oxygen (O) (8p+; 8n0; 8e–) 8e 2e Sodium (Na) (11p+; 12n0; 11e–) Figure 2.6 Formation of an ionic bond. Sodium atom (Na) (11p+; 12n0; 11e–) Chlorine atom (Cl) (17p+; 18n0; 17e–) + – Sodium ion (Na+) Chloride ion (Cl–) Sodium chloride (NaCl) (a) Sodium gains stability by losing one electron, and chlorine becomes stable by gaining one electron. CI– Na+ (c) Large numbers of Na+ and Cl– ions associate to form salt (NaCl) crystals. (b) After electron transfer, the oppositely charged ions formed attract each other. Figure 2.7 Formation of covalent bonds. Reacting atoms Resulting molecules + or Structural formula shows single bonds. Hydrogen atoms Carbon atom Molecule of methane gas (CH4) (a) Formation of four single covalent bonds: Carbon shares four electron pairs with four hydrogen atoms. + or Structural formula shows double bond. Oxygen atom Oxygen atom Molecule of oxygen gas (O2) (b) Formation of a double covalent bond: Two oxygen atoms share two electron pairs. + or Structural formula shows triple bond. Nitrogen atom Nitrogen atom (c) Formation of a triple covalent bond: Two nitrogen atoms share three electron pairs. Molecule of nitrogen gas (N2) Figure 2.8 Carbon dioxide and water molecules have different shapes, as illustrated by molecular models. Figure 2.9 Ionic, polar covalent, and nonpolar covalent bonds compared along a continuum. Figure 2.10 Hydrogen bonding between polar water molecules. + – Hydrogen bond (indicated by dotted line) + + – – – + + + – (a) The slightly positive ends (+) of the water molecules become aligned with the slightly negative ends (–) of other water molecules. (b) A water strider can walk on a pond because of the high surface tension of water, a result of the combined strength of its hydrogen bonds. Figure 2.13 The pH scale and pH values of representative substances. Concentration (moles/liter) Examples [OH–] [H+] pH 100 10–14 14 1M Sodium hydroxide (pH=14) 10–1 10–13 13 Oven cleaner, lye (pH=13.5) 10–2 10–12 12 10–3 10–11 11 10–4 10–10 10 10–5 10–9 9 10–6 10–8 8 10–7 10–7 7 Neutral 10–8 10–6 6 10–9 10–5 5 10–10 10–4 4 10–11 10–3 3 10–12 10–2 2 10–13 10–1 1 10–14 100 0 Household ammonia (pH=10.5–11.5) Household bleach (pH=9.5) Egg white (pH=8) Blood (pH=7.4) Milk (pH=6.3–6.6) Black coffee (pH=5) Wine (pH=2.5–3.5) Lemon juice; gastric juice (pH=2) 1M Hydrochloric acid (pH=0) Figure 2.12 Dissociation of salt in water. + – + Water molecule Salt crystal Ions in solution Figure 2.16c Lipids. (c) Simplified structure of a steroid Four interlocking hydrocarbon rings form a steroid. Example Cholesterol (cholesterol is the basis for all steroids formed in the body) Figure 2.19 Levels of protein structure. Amino acid Amino acid Amino acid Amino acid Amino acid (a) Primary structure: The sequence of amino acids forms the polypeptide chain. (b) Secondary structure: The primary chain forms spirals (-helices) and sheets (-sheets). -Helix: The primary chain is coiled to form a spiral structure, which is stabilized by hydrogen bonds. -Sheet: The primary chain “zig-zags” back and forth forming a “pleated” sheet. Adjacent strands are held together by hydrogen bonds. (c) Tertiary structure: Superimposed on secondary structure. -Helices and/or -sheets are folded up to form a compact globular molecule held together by intramolecular bonds. (d) Quaternary structure: Two or more polypeptide chains, each with its own tertiary structure, combine to form a functional protein. Tertiary structure of prealbumin (transthyretin), a protein that transports the thyroid hormone thyroxine in serum and cerebrospinal fluid. Quaternary structure of a functional prealbumin molecule. Two identical prealbumin subunits join head to tail to form the dimer. Figure 2.19a Levels of protein structure. Amino acid Amino acid Amino acid Amino acid Amino acid (a) Primary structure: The sequence of amino acids forms the polypeptide chain. Figure 2.19b Levels of protein structure. -Helix: The primary chain is coiled to form a spiral structure, which is stabilized by hydrogen bonds. -Sheet: The primary chain “zig-zags” back and forth forming a “pleated” sheet. Adjacent strands are held together by hydrogen bonds. (b) Secondary structure: The primary chain forms spirals (-helices) and sheets (-sheets). Figure 2.19c Levels of protein structure. Tertiary structure of prealbumin (transthyretin), a protein that transports the thyroid hormone thyroxine in serum and cerebrospinal fluid. (c) Tertiary structure: Superimposed on secondary structure. -Helices and/or -sheets are folded up to form a compact globular molecule held together by intramolecular bonds. Figure 2.19d Levels of protein structure. Quaternary structure of a functional prealbumin molecule. Two identical prealbumin subunits join head to tail to form the dimer. (d) Quaternary structure: Two or more polypeptide chains, each with its own tertiary structure, combine to form a functional protein. An example of the progression in complexity of structure in proteins with the final quaternary structure being that of hemoglobin. Figure 2.20 Enzymes lower the activation energy required for a reaction to proceed rapidly. WITHOUT ENZYME WITH ENZYME Activation energy required Less activation energy required Reactants Reactants Product Product Figure 2.21 Mechanism of enzyme action. Substrates (S) e.g., amino acids + Product (P) e.g., dipeptide Energy is absorbed; bond is formed. Water is released. Peptide bond Active site Enzyme (E) Enzyme-substrate complex (E-S) 1 Substrates bind 2 Internal at active site. rearrangements Enzyme changes leading to shape to hold catalysis substrates in occur. proper position. Enzyme (E) 3 Product is released. Enzyme returns to original shape and is available to catalyze another reaction. Substrate “fits” with active site Substrate unable to bind Active site Denatured enzyme Functional enzyme (a) (b) Active site Amino acids + Enzyme (E) Substrates (S) Enzyme-substrate complex (E-S) H2O Free enzyme (E) Peptide bond Internal rearrangements leading to catalysis Dipeptide product (P) Figure 2.22 Structure of DNA. Phosphate Sugar: Deoxyribose Base: Adenine (A) Thymine (T) Adenine nucleotide Sugar Phosphate Thymine nucleotide Hydrogen bond (a) Sugar-phosphate backbone Deoxyribose sugar Phosphate Adenine (A) Thymine (T) Cytosine (C) Guanine (G) (b) (c) Computer-generated image of a DNA molecule Figure 2.23 Structure of ATP (adenosine triphosphate). High-energy phosphate bonds can be hydrolyzed to release energy. Adenine Phosphate groups Ribose Adenosine Adenosine monophosphate (AMP) Adenosine diphosphate (ADP) Adenosine triphosphate (ATP) Figure 2.24 Three examples of cellular work driven by energy from ATP. Solute + Membrane protein (a) Transport work: ATP phosphorylates transport proteins, activating them to transport solutes (ions, for example) across cell membranes. + Relaxed smooth muscle cell Contracted smooth muscle cell (b) Mechanical work: ATP phosphorylates contractile proteins in muscle cells so the cells can shorten. + (c) Chemical work: ATP phosphorylates key reactants, providing energy to drive energy-absorbing chemical reactions. Table 2.1 Common Elements Composing the Human Body (1 of 2) Notice how there are three broad categories of these elements, major, lessor, and trace. Table 2.1 Common Elements Composing the Human Body (2 of 2)