* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Day 1. General aspects for genetic map construction
Survey
Document related concepts
Transcript
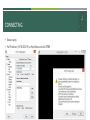
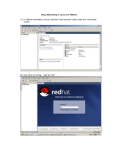
DAY 1. GENERAL ASPECTS FOR GENETIC MAP CONSTRUCTION SANGREA SHIM INDEX Day 1 General aspects for genetic map construction Genetic polymorphism and recombination frequency Genotyping using molecular marker Map construction (phenotype, AFLP, RFLP) Sequencing method Next generation sequencing Whole genome reference sequence Reference sequencing for Genotyping Retrieving sequence polymorphism Genetic map construction (SNP, InDel) GENETIC POLYMORPHISM & RECOMBINATION FREQUENCY GENOTYPING USING MOLECULAR MARKER An Integrated High-density Linkage Map of Soybean with RFLP, SSR, STS, and AFLP Markers Using A Single F2 Population Xia et al. 2008 MAP CONSTRUCTION An Integrated High-density Linkage Map of Soybean with RFLP, SSR, STS, and AFLP Markers Using A Single F2 Population Xia et al. 2008 NEXT GENERATION SEQUENCING Sequencing Sanger’s Dideoxy Termination Using dNTPs Electrophoresis in capillary gel Read dye colors one-by-one Average 700~900 bp Massive Parallel Sequencing Platform So called Next Generation Sequencing platform SOLiD (Sequencing by Ligation), Illumina (Sequencing by synthesis), 454 (Pyrosequencing) Read 50+35(50+50), 50~300, 700 bp 1200~1300, ~3000, 1 million reads per run NEXT GENERATION SEQUENCING Sequencing technologies – the next generation Michael et al. Nature review genetics 2010 WHOLE GENOME REFERENCE SEQUENCE Polymorphism discovered by comparison Reference is required for comparison So, the reference genome is obligated Making contigs which is constituted by unique sequences combination using PE or small size MP Scaffolding which includes less unique sequences (i.e. repetitive sequences) using large insert size MP library sequences Anchor the scaffold using genetic map But, genetic map constituted by several types of molecular marker is not able to translate to sequence information RESEQUENCING FOR GENOTYPING GET Polymorphism!, Treat it as a marker or locus! SNPs Small size InDels Align several depth of raw read sequence against Ref. Statistics Lots of alignment software is available BLAST, BLAT, BWA, BOWTIE-series….. Aligner which use BWT as a main algorithm are famous Fast, efficient RESEQUENCING FLOW CHART DNA/RNA NGS platform Alignment pileup bwa bowtie2 samtools bcftools VCF Raw read Sequences SAM samtools selection BAM Quality trimming SolexaQA samtools Sorted BAM Map construction JoinMap4 RETRIEVING SEQUENCE POLYMORPHISM BOWTIE2 or BWA are just align the bulky reads to reference sequence Making SAM(sequence alignment/mapping)/BAM(binary sequence alignment/mapping) as a result Several types of statistics or inferences can be adapted to retrieving polymorphism (Picard, GATK) Samtools package is used in retrieving variants The VCF(variant calling format) is the ouput file GENETIC MAP CONSTRUCTION Selection of a core set of RILs from Forrest x Williams 82 to develop a framework map in soybean Wu et al. 2011 HURDLES ON THE ROAD TO GENETIC MAP Output of calling variation is a VCF format JoinMap input file is LOC format Is there a Converter between the VCF and LOC? Make converter program, Make genetic map yourself These are the final goal of this courses TODAY’S PRACTICE Make a connection to remote computer Get used to Linux system Get familiar with python2.7 THANK YOU If you have a question, please ask me. DAY 1. PRACTICE - BASIC LINUX COMMAND TAEYOUNG LEE CONNECTING Server is located in Seoul National University campus Connect to server computer using putty SSH client program Download at http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html CONNECTING Execute putty Put IP address (147.46.250.193) at Host Name and click OPEN CONNECTING ID : trainee PW : bogor Then you are in server now Only white character on black background BASIC COMMAND IN LINUX ls Listing files and directories cd Change directory Practice) enter into /data2/python BASIC COMMAND IN LINUX mkdir Make directory Usage) mkdir dir_name Practice) make directory named as your name BASIC COMMAND IN LINUX vi Open text editing program Make new text file usage) vi filename_to_edit vi filename_to_make Practice) make text file named as yourname in your directory, write something and save it Insert, replace, esc :q :w :wq :q! BASIC COMMAND IN LINUX mv Moving files or directories Rename files or directories Usage) mv present_file_path file_path_to_move Practice) Change directory into upper directory cm) cd .. Make some text file by vi Move text file to your directory Rename text file BASIC COMMAND IN LINUX cp Coping files or directories Usage) cp file_path file_path_to_copy cp can rename file If you want to copy directory, you have to use –r option Cp –r dir_path dir_path_to_copy Practice) Make directory in your directory Copy some file into directory with rename and w/o rename BASIC COMMAND IN LINUX rm Removing files or directories Usage) rm file_name If you want to remove directory, you have to use –r option rm –r dir_name Practice) Remove the directory and file BASIC COMMAND IN LINUX less Read only text viewer Have advantage for large size text file Usage) less file_name Searching function / Practice) Open large text file by vi and less /data2/python/Gmax_109_gene_exons.gff3 Use searching function /Gm12 wget ftp://ftp.arabidopsis.org/ home/tair/Sequences/whole_chromosomes/tai r9_Assembly_gaps.gff BASIC COMMAND IN LINUX cat Concatenate files Print out files Usage cat file_name1 file_name2 … Practice) Print out file by cat Print out file three times BASIC COMMAND IN LINUX grep Grep the lines contain some words Usually use with cat Usage) cat file_name | grep ‘word’ ‘|’ mean after This usage mean we grep line which contain some word after print out file Various useful options -v : vanish -c : count ‘word1\|word2’ = word1 or word2 grep ‘word1’ | grep ‘word2’ = word1 and word2 Practice) Grep ‘Gm12’ in /data2/python/Gmax_109_gene_exons.gff3 Grep ‘Gm12’ or ‘Gm15’ in same file Grep ‘gene’ and ‘mRNA’ Count line contain ‘Gm12’ Vanish line contain exon or CDS or mRNA BASIC COMMAND IN LINUX sort Sorting file Usually use with cat Usage) cat file_name | sort Various useful options -k sort by column -u sort and remove redundancy -n numeric sort -r reverse -d delimiter setting Practice) Sort /data2/python/Gmax_109_gene_exons.gff3 by start position(by column and numeric) BASIC COMMAND IN LINUX cut Cutting column in file Usually use with cat Usage) cat file_name | cut –f n (n : integer) Practice) Retrieve chromosome, start position, end position in /data2/python_study/Gmax_109_gene_exons.gff3 BASIC COMMAND IN LINUX > Standard input, output vs. file input, output Input and output on screen or file > can save standard output to file output cat file_name | grep ‘word’ > output_file >> >> also can save standard output to file output But just adding! HANDLE FILE Fasta file /data2/python/ap2.fa Fastq file /data2/python/example.fastq Gff file /data2/python/Gmax_109_gene_exons.gff3 Python file! /data2/python/1stday.py Make a new text file named as new.txt The file contain Gm01,1,23 Gm04,4,56 Gm03,6,78 Gm04,8,10 Copy new.txt into new.copy Remove new.copy Using cat, print the contents of new.txt Using grep, print the contents the new.txt contain Gm04 Using cut, print the first column of new.txt and save it as a file named as new.txt.cut THAT’S IT FOR TODAY Q &A