* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PLPD: Protein Localization Prediction for Imbalanced and
Survey
Document related concepts
Vectors in gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene nomenclature wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcript
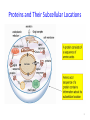
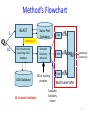
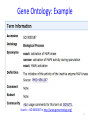
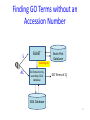
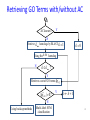
Semantic Similarity over Gene Ontology for Multi-label Protein Subcellular Localization Shibiao WAN and Man-Wai MAK The Hong Kong Polytechnic University Sun-Yuan KUNG Princeton University Outline 1. 2. 3. 4. 5. 6. Introduction and Motivation Retrieval of GO Terms Semantic Similarity Measures Multi-label Multi-Class Classification Results Conclusions 2 Proteins and Their Subcellular Locations 3 Subcellular Localization Prediction • The subcellular locations of proteins help biologists to elucidate the functions of proteins. • Identifying the subcellular locations by entirely experimental means is time-consuming and costly. • Computational methods are necessary for subcellular localization prediction. • Previous research has found that gene ontology (GO) based methods outperform methods based on other protein features (e.g. AA composition). 4 Multi-label Problem • Some proteins can simultaneously reside at, or move between, two or more subcellular locations. • Multi-label (Multi-location) proteins play important roles in some metabolic processes taking place in multiple subcellular locations. • State-of-the-art multi-label predictors, such as PlantmPLoc, iLoc-Plant, and mGOASVM use frequency counts of GO terms as features. • In this work, we propose using semantic similarity of GO terms as features for multi-label subcellular localization prediction. 5 Method’s Flowchart S BLAST Swiss-Prot Database SVM GO Extraction by searching GOA database Semantic Similarity Measure SVM GOA Database GO of training proteins . . . homolog AC AC SS: Semantic Similarity M . . . Subcellular Location(s) SVM Multi-label SVM Semantic Similarity Vector 6 Gene Ontology Gene ontology is a set of standardized vocabularies annotating the functions of genes and gene products GO terms, e.g., GO:0000187 A protein sequence may correspond to 0, 1 or many GO terms. 7 Gene Ontology: Example Search----GO:0000187 in http://www.geneontology.org/ 8 GOA Database • Gene Ontology Annotation database. – Provide structured annotations to proteins in UniProt Knowledgebase (UniProtKB) and other protein databases using standardized GO vocabularies. – Include a series of cross-references to other databases. • Given an Accession Number, the GOA database allows us to find a set of GO terms associated with that accession number. 9 GOA Database Accession Number (AC) GO term(s) Search A0M8T9 in http://www.ebi.ac.uk/GOA/ 1 AC maps to many GO terms ! 10 Finding GO Terms without an Accession Number S BLAST Swiss-Prot Database homolog AC AC GO Extraction by searching GOA database GO Terms of Qi GOA Database 11 Semantic Similarity Measure GO term x Find Common Ancestors A(x,y) GO term y Ancestors Computing Semantic Similarity sim(x,y) SQL Query GO Database 12 Finding Common Ancestors, A(x,y) 13 Finding Common Ancestors, A(x,y) GO:0000187 is_a part_of 14 Semantic Similarity Measure We use Lin’s measure to estimate the semantic similarity between two GO terms (x and y): p(c) = #(proteins annotated to GO term c) #(all proteins annotated to the GO taxonomy) 15 Semantic Similarity between 2 Proteins Semantic similarity between 2 proteins (Gi, Gj): where Semantic Similarity Vector: No. of training proteins 16 Multi-label SVM Scoring GO of Qt GO of training proteins = 17 Benchmark Datasets The Plant dataset 18 Performance Metrics Overall locative accuracy: Overall actual accuracy: Actual accuracy is more objective and stricter! 19 Performance Comparison The Plant dataset 20 Conclusions • Our Proposed predictor performs significantly better than Plant-mPLoc and iLoc-Plant, and also better than mGOASVM, in terms of locative and actual accuracies. • As for individual locative accuracies, our proposed predictor are significantly higher than the three predictors for all of the 12 locations. • In terms of GO information extraction, Plant-mPLoc, iLoc-Plant and mGOASVM use the occurrences of GO terms as features, whereas the proposed predictor discovers the semantic relationship between GO terms, from which the semantic similarity between proteins can be obtained. 21 Web Servers 22 Thank you! 23 Multi-label SVM Classifier Transformed labels for M-class problem: 24 Retrieving GO Terms with/without AC AC known ? Y N Retrieve k max homologs by BLAST;k 1 k 0 Using the k - th homolog N k kmax ? Y Retrieve a set of GO terms G i,ki |G i ,k i | 0 ? Using back-up methods N Multi-label SVM classification Y k k 1 25 Finding Common Ancestors • The relationships between GO terms in the GO hierarchy can be obtained from the SQL database through the link: http://archive.geneontology.org/latesttermdb/go_daily-termdb-tables.tar.gz. • We only considered the ‘is-a’ relationship. 26