* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Programming Biological Cells - MIT Computer Science and Artificial
Survey
Document related concepts
Transcript
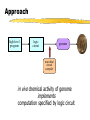
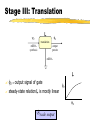
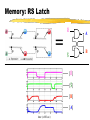
Toward in vivo Digital Circuits Ron Weiss, George Homsy, Tom Knight MIT Artificial Intelligence Laboratory Motivation Goal: program biological cells Characteristics small (E.coli: 1x2m , 109/ml) self replicating energy efficient Potential applications “smart” drugs / medicine agriculture embedded systems Approach high-level program logic circuit genome microbial circuit compiler in vivo chemical activity of genome implements computation specified by logic circuit Key: Biological Inverters Propose to build inverters in individual cells each cell has a (complex) digital circuit built from inverters In digital circuit: signal = protein synthesis rate computation = protein production + decay Digital Circuits With these inverters, any (finite) digital circuit can be built! A A B C D = C D gene C B gene proteins are the wires, genes are the gates NAND gate = “wire-OR” of two genes gene Outline Compute Model using Inversion and Simulations Measuring Microbial Related signals and circuits Circuit Design work Conclusions & Future Work Components of Inversion Use existing in vivo biochemical mechanisms stage I: cooperative binding found in many genetic regulatory networks stage II: transcription stage III: translation decay of proteins (stage I) & mRNA (stage III) examine the steady-state characteristics of each stage to understand how to design gates Stage I: Cooperative Binding fA input protein C cooperative binding rA repression input protein C fA = input protein synthesis rate rA = repression activity rA (concentration of bound operator) steady-state relation C is sigmoidal “clean” digital signal 0 fA 1 Stage II: Transcription T rA yZ transcription repression mRNA synthesis rA = repression activity yZ = mRNA synthesis rate steady-state relation T is inverse invert signal T yZ rA Stage III: Translation L yZ fZ translation mRNA synthesis output protein mRNA fZ = output signal of gate steady-state relation L is mostly linear L fZ yZ scale output Putting it together fA input protein C cooperative binding rA T transcription repression yZ L mRNA synthesis output protein mRNA inversion relation I : fZ = I (fA) = L ∘ T fZ translation input protein signal I ∘ C (fA) “ideal” transfer curve: gain (flat,steep,flat) adequate noise margins “gain” fZ 0 fA 1 Outline Compute Model using Inversion and Simulations model based on phage steady-state and dynamic behavior of an inverter simulations of gate connectivity, storage Measuring Microbial Related signals and circuits Circuit Design work Conclusions & Future Work Model Understand Model general characteristics of inversion phage elements [Hendrix83, Ptashne92] repressor (CI) operator (OR1:OR2) promoter (PR) output protein (dimerize/decay like CI) OR2 OR 1 structural gene [Ptashne92] Steady-State Behavior Simulated transfer curves: fA fB fA fB Title: inv erter-eqlb-for-dimacs 99-talk.eps Creator: gnuplot 3.5 (pre 3.6) patc hlev el beta 340 Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. fC Title: tw o-inv erters -eqlb-for-dimacs 99-talk.eps Creator: gnuplot 3.5 (pre 3.6) patc hlev el beta 340 Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. fA fB fA asymmetric (hypersensitive to LOW inputs) later in talk: ways to fix asymmetry, measure noise margins fC Inverter’s Dynamic Behavior Dynamic behavior shows switching times [A] [ active gene ] [Z] time (x100 sec) Connect: Ring Oscillator Connected gates show oscillation, phase shift [A] [B] [C] time (x100 sec) Memory: RS Latch _ R = A _ S B _ [R] _ [S] [B] [A] time (x100 sec) Outline Compute Model using Inversion and Simulations Measuring signals and circuits measure a signal approximate a transfer curve (with points) the transfer band for measuring fluctuations Microbial Related Circuit Design work Conclusions & Future Work Measuring a Signal Attach a reporter to structural gene Translation phase reveals signal: n copies of output protein Z m copies of reporter protein RP (e.g. GFP) Signal: Time derivative: Title: meas ure-signal-eqn-3.dv i Creator: dvips (k) 5.78 Copyright 1998 Radic al Eye Softw are (w w w .radicaleye.c om) Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Ti tle: Comment: measure-si This EPS picture w ill print to gnal-eqn-2.dvi a Pos tSc ript printer,Creator: but not to dvi ps(k) 5.78 Copyright 1998 Radi cal Eye Software (www.radical eye.com) other ty pes of printers. Preview: Title: This EPS picture was not saved measure-signal-eqn-4.dvi wi th a previ ew included in it. Creator: Comment: dvips(k) 5.78 Copyright 1998 will Radical Title: This EPS picture printEye to aSoftw are (w w w .radicaleye.com) Preview : PostScri pt pri nter, but not to measure-signal-eqn-8.dvi This EP S picture as not otherw types of saved pri nters. Creator: w ith a preview included in it. 1998 Radical Eye Software (ww w.radicaleye.com) dvips(k) 5.78 Copyright Comment: Title: Preview: This EP S picture w ill print a saved measure-signal-eqn-7.dvi This EPS picture wastonot [in equlibrium] Measured signal: Measuring a Transfer Curve To measure a point on the transfer curve of an inverter I (input A, output Z): Construct a “fixed drive” (with reporter) a constitutive promoter with output protein A measure reporter signal fA A RP Z RP “drive” gene Construct “fixed drive” + I (with reporter) measure reporter signal fZ “drive” gene Result: A inverter point (fA, fZ) on transfer curve of I Measuring a Transfer Curve II Approximate the transfer curve with many points Example: • 3 different drives • each with cistron counts 1 to 10 Title: meas ure-tf-w ith-points-talk.eps Creator: gnuplot 3.5 (pre 3.6) patc hlev el beta 340 Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. fZ fA mechanism also useful for more complex circuits Models vs. Reality Need to measure fluctuations in signals Use flow cytometry get distribution of fluoresence values for many cells Title: cell suspension single-cell luminosity readout Creator: gnuplot Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. typical histogram of scaled luminosities for “identical” cells The Transfer Band The transfer band: captures systematic fluctuations in signals constructed from dominant peaks in histograms For histogram peak: output min/max = fA/fA Each pair of drive + inverter signals yield a rectangular region fZ fZ fA fA input Outline Compute Model using Inversion and Simulations Measuring Microbial signals and circuits Circuit Design issues in building a circuit matching gates modifying gates to assemble a library of gates BioSpice Related work Conclusions & Future Work Microbial Circuit Design Problem: Need gates have varying characteristics to (1) measure gates and construct database (2) attempt to match gates (3) modify behavior of gates (4) measure, add to database, try matching again Simulate & verify circuits before implementing Matching Gates Need to match gates according to thresholds output HIGH Imax Imin Imin(Iil) Imax(Iih) LOW Iil LOW Iih Title: match-gates-eqn-1.dvi Creator: dvips (k) 5.78 Copyright 1998 Radic al Eye Softw are (w w w .radic aley e.c om) Prev iew : This EPS picture w as not s av ed Title: w ith a preview inc luded in it. match-gates-eqn-2.dvi Comment: Creator: This EPS picture w ill print to a dvips (k) 5.78 Copyright 1998 Radic al Eye Softw are (w w w .rad Pos tSc ript printer, but not to Prev iew : other ty pes of printers. This EPS Title: picture w as not s av ed w ith a preview inc luded in it. match-gates -eqn-3.dvi Comment:Creator: This EPS dv picture ill print to a ips(k)w5.78 Copyright 1998 Radical Ey e Softw are ( Pos tSc ript printer, but not to Preview : input other ty pes printers. Thisof EPS picture w as not saved w ith a preview included in it. HIGH Comment: Modifications to Gates modification stage Modify repressor/operator affinity C Modify the promoter strength T Alter degradation rate of a protein Modify RBS strength Increase Add cistron count autorepression C L T C Each modification adds an element to the Modifying Repression Reduce repressor/operator binding affinity use base-pair substitutions Schematic effect on cooperative-binding stage: Simulated effect on entire transfer curve: C fZ rA Title: inv erter-eqlb-k_rprs-for-dimac s99-talk.eps Creator: gnuplot 3.5 (pre 3.6) patc hlev el beta 340 Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. fA fA Modifying Promoter Reduce RNAp affinity to promoter Schematic effect on transcription stage: Simulated effect on entire transfer curve: T yZ fZ Title: inv erter-eqlb-k_prom-dimacs 99-talk.eps Creator: gnuplot 3.5 (pre 3.6) patc hlev el beta 340 Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. rA fA BioSpice Prototype simulation & verification tool intracellular circuits, intercellular communication Given a circuit (with proteins specified) simulate concentrations/synthesis rates Example circuit to simulate: messaging + setting state BioSpice Simulation Small colony: 4x4 grid, 2 cells (outlined) (1) original I=0 (2) introduce D send msg M (3) recv msg set I (4) msg decays I latched Limits to Circuit Complexity amount of extracellular DNA that can be inserted into cells reduction in cell viability due to extra metabolic requirements selective pressures against cells performing computation probably not: different suitable proteins Related Work Universal automata with bistable chemical reactions [Roessler74,Hjelmfelt91] Mathematical models of genetic regulatory systems [Arkin94,McAdams97,Neidhart92] Boolean networks to describe genetic regulatory systems [Monod61,Sugita63,Kauffman71,Thomas92] Modifications to genetic systems [Draper92, vonHippel92,Pakula89] Conclusions + Future Work in vivo digital gates are plausible Now: Implement and measure digital gates in E. coli Also: Analyze robustness/sensitivity of gates Construct a reaction kinetics database Later: Study proteinprotein interactions for faster circuits Inverter: Chemical Reactions Title: reac tions -table.dvi Creator: dvips k 5.58f Copyright 1986, 1994 Radic al Ey e Softw are Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers. Title: kinetic -rates.dv i Creator: dvips k 5.58f Copyright 1986, 1994 Radical Ey e Prev iew : This EPS picture w as not s av ed w ith a preview inc luded in it. Comment: This EPS picture w ill print to a Pos tSc ript printer, but not to other ty pes of printers.