* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Evolutionary quantitative genetics and one
Viral phylodynamics wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Medical genetics wikipedia , lookup
Genetic drift wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Selective breeding wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Koinophilia wikipedia , lookup
Group selection wikipedia , lookup
Heritability of IQ wikipedia , lookup
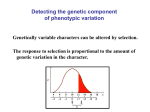
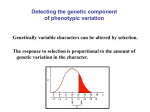
“Coarse” Notes Population Genetics Evolutionary quantitative genetics and one-locus population genetics READING: Hedrick pp. 57–63, 587–596 • Most evolutionary problems involve questions about phenotypic means • Goal: determine how selection causes evolutionary change in the mean of a quantitative character. • Aside: Quantitative vs. Qualitative Traits • Back to our main story.... selection on a trait such as body size – body size of individual is denoted z – mean body size in a population: z – Suppose selection alters allele frequencies at loci that affect z – Consider the effect on body size of one specific locus, i – In general, to determine how much z changes, one needs to determine: 1) how selection changes p (i ) at all loci affecting z 2) how changes in p (i ) combine to change z . body size z Δz ( i) ( i) Δp (i ) 0 p (i ) – We'll focus on the effects of one of the loci that affects z • The phenotype of an individual depends on: 1) its genotype at locus i 2) its genotype at other loci that affect the trait 3) its environmental experience VI-1 1 “Coarse” Notes Population Genetics • lump 2 & 3 into a single factor, c • Can think of an individual's phenotype as z = a jk + c a22 z =c+ a12 a 11 A1 A1 A1 A2 A2 A2 • The average phenotype can similarly be broken down into the average contribution of locus i and the average over other loci and environmental effects: z = a + c • Consider the effect of a change in p (i ) on z (holding all other loci constant) !z ( i) (drop i) ( ) ( ) = z p( i) + !p( i) " z p( i) ≈ dz (i ) (i ) !p dp ≈ % " d ln w ' dz $ 1 (term in brackets is just our old friend) 2 p(1 ! p) 24 3 dp ' dp $ 14 variance 123 # selection & ≈ dz " 1 dz d ln w % (using the "chain rule") 2 p(1 ! p) dp $# dp dz '& ≈ 1 2 2 or !z ( i) ! dz $ d ln w pq# " dp % dz (form emphasizes the dependency of fitness w on phenotype z) – What is dz ? dp VI-2 “Coarse” Notes Population Genetics dz d (a + c ) = dp dp d 2 = p a1 1 + 2pqa1 2 + q 2 a2 2 + c ) (assuming H- W) ( dp = 2pa1 1 + 2qa1 2 ! 2 pa1 2 ! 2qa2 2 + 0 – So, putting all the pieces together: !z ( i) " 12 # pq # 4( pa1 1 + qa1 2 $ pa1 2 $ qa2 2) 2 d ln w dz or !z ( i) " G( i) # (i ) where G = 2pq[( pa1 1 + qa1 2 ) ! ( pa1 2 + qa2 2 )] 2 and ! = d ln w . dz – What is ! ? • Called the selection gradient • It is a measure of the strength of selection acting on a trait: (i ) – What is G ? • It is an extremely important quantity called the additive genetic variance contributed by locus i • Why "additive"? – Dominance Variance • Consider the total amount of variation in z due to variation at locus i: ( ) var z( i) = var ( ajk + c ) = var ( ajk ) since we have fixed c. ( ) – So, var z( i) = (assuming H-W equilibrium) = var ( ajk ) (p a 2 2 11 [ ] E( a jk ) ! E( a jk ) 2 = 2 + 2 pqa1 2 + q 2 a2 2) ! ( p2 a1 1 + 2pqa1 2 + q 2 a2 2) 2 VI-3 2 2 “Coarse” Notes Population Genetics (after a lot of tedious algebra) = = G (i ) a + a $) & " + '2 pq a1 2 ! 1 1 2 2 * # 2 %+ ( 2 G( i ) + D(i ) 2 a + a $) & " where D = '2 pq a1 2 ! 1 1 2 2 * . # 2 %+ ( (i ) (i ) • What is D ? – It's called the dominance variance contributed by locus i. (i ) • Note: D is never negative and has units of z2 – Why "dominance"? (i ) (i ) – Relationship between G and D • Case 1: No Dominance • Case 2: Symmetric Overdominance – What about other loci that contribute to the traits? • Generally difficult (recall complications of 2-locus population genetics) to understand; • Assuming linkage equilibrium for all loci contributing to a trait: !z = !z (1) + !z ( 2 ) + !z( 3 ) +L [ ] = G(1 ) + G (2 ) + G (3 ) +L " " n (i ) % = $! G '( or # i =1 & !z = G " VI-4 (recall that ! reflects how a change in mean phenotype changes mean population fitness: it doesn't care what the cause of the change in phenotype is.) “Coarse” Notes Population Genetics – This is the central equation of quantitative genetics – Note its similarity to the equation for selection at one locus. – Note that it's written in several other ways • Important Message: It's not the total phenotypic variance nor even the total genotypic variance that determines how fast a population mean evolve in response to selection n – Total genotypic variance = G + D (where D = D( 1) + D( 2 ) +L= ! D( i) ) i =1 • In general, there's between-locus genetic "interaction" variance as well. – Total phenotypic variance = G + D + E (also written V P = V A + V D + V E ) • E is the variance that's due to environmental effects. • A few quibbles with “heritability” Selection on a single character ( !z = G " ): Other ways of determining G • We now have an algebraic definition of G in terms of a jk ' s and p's at all loci • There are, however, other ways to get G – We'll skip the tedious algebra (see Falconer), but the methods are important since they show how G can actually be measured! (most of Falconer's book concerns estimating G in agricultural or laboratory settings. See, e.g., Simms & Rausher for methods of measuring G in natural populations) (1) Breeding Values – Measure the type of offspring an individual tends to produce in a particular reference population • breeding values were originally used by plant and animal breeders for assessing the value of an individuals for mating. – Definition: g = z + 2(zo ! z ) VI-5 “Coarse” Notes Population Genetics • note that an individual's own phenotype doesn't enter in this - except through z • zo = average value of the trait among the individual's offspring when she/he is mated to a large # of randomly chosen individuals – alternatively, zo may be thought of as the expected phenotype the individual's offspring were it to mate with a randomly chosen individual in the population. • Why the 2? – Only 1/2 of an offspring phenotype is attributable to that individual. The other 1/2 comes from its mate. • Important note: the breeding value of an individual refers to a particular population as well as the individual. – That is, the same individual could have a different breeding value were it moved to a different population. • Uses of breeding value – Suppose a farmer (or nature) chooses a set of individuals to reproduce – The mean value of the offspring will equal the average breeding value of the selected individuals • Breeding values can be estimated – In principle, one could 1) mate an individual to many others 2) raise and measure the offspring 3) estimate the breeding value using the average offspring phenotype • This is actually done for important breeding stock, dairy cattle, race horses, etc. – What about variation in breeding values? • It's obviously related to a population's genetic variation • can actually compute it under our earlier assumptions • It turns out that var ( g) = G VI-6 “Coarse” Notes Population Genetics – Significance • Statistically, can think of the phenotype of an individual as z = g + d + e – g and d are inherited from its parents, but the individual will only pass on g – By the way g and d are defined, d is acquired by an offspring independently of g • These results suggest a way to measure G without ever knowing the p's and aij's! – could experimentally determine breeding values of individuals in a population, then compute their variance – this is, essentially, what a "half-sib" breeding design does – Drawback: this method is rather tedious, expensive, and labor intensive • Turns out there's an easier way... (2) Parent-offspring regression – The response to selection, "z , depends on the relationship between parents and offspring diagrammed here. – "z depends on ! ! • how strongly selection acts on the mean (s) • the orientation of the cloud of points – note that the orientation depends on inheritance • e.g., if there's no genetic variance, the regression line would be flat. – This orientation is related to genetics. It can be show (see H&C or Falconer) that, under our previous assumptions: G = 2cov ( zo ,zp ) – This is measurable (and, once again, doesn't require estimating the p's and aij's). – It's one of the major methods for estimating G Selection on a single character ( !z = G " ): The selection gradient revisited • We'll now consider two definitions of the selection gradient VI-7 “Coarse” Notes Population Genetics (1) ! = change in the logarithm of a population's mean fitness that would result if the population's mean phenotype was increased by one "unit" • Mathematically, ln[ w (z + ! )] " ln[ w (z )] # i.e., ! = d ln w ln w ( z + # ) $ ln w (z ) " # dz d ln w ! = $! , dz • ! measures steepness of mean population fitness function – this is the relevant measure of the strength of selection acting on the mean. • The units of ! are 1/char. • The relationship between w (z) , w , and ! : – w (z) : individual fitness function • describes the fitness of an individual as a function of it's phenotype – w : population mean fitness • it's the average fitness of individuals in the population • i.e., it's the average of w (z) with respect to the distribution of phenotypes z in the population – The relationship between w (z) and w : • Suppose that ecologists/evolutionary biologists determine the following fitness function a) If P = 0 (i.e., there is no phenotypic variation => all individuals are identical) then, w = w( z) = w(z ) since z = z . – Note: w = w ( z ) : b) If there's lots of phenotypic variation (P >> 0): • In general, w is, roughly, a smooth version of w (z) . • "weak selection": – ! is approximately independent of P VI-8 “Coarse” Notes Population Genetics • or, w (z) changes little over the distribution of z – refers to stabilizing or disruptive component of selection – note that the directional component of selection may still be strong (2) ! = regression of relative fitness onto the value of the character • Mathematically, ! = cov ( w, z) var (z ) – this indicates how ! can be measured! Effects of selection within a generation: the "selection differential" • The selection differential measures the within-generation impact of selection on a population's mean phenotype. * • Mathematically, the selection differential is defined as s = z ! z • Another mathematically equivalent definition (due to Robertson & Price): s = cov (w, z) • The relationship between the selection gradient and selection differential: s = P! !=s P – Note: for a single trait, there seems little difference between s and ! . However, when the simultaneous evolution of several quantitative traits is being considered, the selection gradient have many advantages over the selection differential (both mathematically and in biological interpretation). • Traditional form for the response to selection: !z = G " = G s G G 2 2 is the "narrow-sense" heritability of z. = s = h s where h = P P P – Comments on heritability: • There are two types: "broad-sense" and "narrow-sense" – Broad-sense heritability is the fraction of phenotypic variability that is due to all forms genetic variation = (G + D + "epistatic variance")/P. VI-9 “Coarse” Notes Population Genetics 2 – Narrow-sense heritability ( h ) is the fraction of phenotypic variability due to additive-genetic variance. 2 • h and s are relatively uninformative regarding rates of evolution (compared to G and ! ): – without changing G and ! (and hence the rate of evolution !z = G " ), one can 2 ↑ s simply by ↑ P (=> h ↓). • Breeders like s because it is easier to control. Evolutionary Equilibrium under selection: !z = G " • !z = 0 for two possible reasons: (1) ! = 0 (2) G = 0. – Possibility (1) corresponds to a balance of ecological forces • ˆz (the equilibrium mean of the population) is either a maximum of minimum of w – the former is stable, the latter is unstable • If selection is sufficiently weak, z will evolve in such a way that mean fitness will ↑. – Note: fitness refers to overall fitness, not to antagonistic components of selection that may occur within a generation e.g., Darwin's finches: Price found that small size is favored in juveniles (possibly due to energy conservation) but large size is favored in adults (easier to crack seeds). – Possibility (2) corresponds to a lack of evolutionary "material" • This situation correponds to what are commonly called "developmental" or "genetic" constraints. • additive variance can reappear in several ways – mutation & migration VI-10 “Coarse” Notes Population Genetics – recombination between loci • linkage disequilibrium affects the amount of variance expressed. e.g., a population with only (+ -) (- +) gametes has no phenotypic variance despite having copious amounts of genetic variance. – recombination can create additive genetic variance – recombination can destroy variation as well (e.g., (+ +) (- -) population.) VI-11