* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download bio98a_l09
Survey
Document related concepts
Transcript
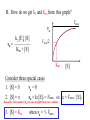
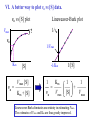
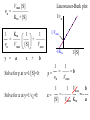
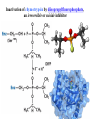
Bio 98 - Lecture 9 Enzymes II: Enzyme Kinetics Amino acid side chains with titratable groups Appr. pKa Carboxylate 4 Amine/guanidi nium 10-12 Sulfhydryl 8 Imidazole 6 Hydroxyl 13 (lecture 8!) Hydroxyl 10 Enzyme catalyzed reaction G (free energy) S E P S‡ ES‡ G‡ E+S E+P G Reaction coordinate 1. Enzymes do not alter the equilibrium or G. 2. They accelerate reactions by decreasing G‡. 3. They accomplish this by stabilizing the transition state. I. Enzyme reactions have at least two steps E+S k1 k-1 binding step - rapid - reversible ES k2 E+P catalytic step - slower - irreversible (often) ES = “enzyme-substrate complex” ≠ transition state (ES‡) (1) What is the physical meaning of the constants? What do they tell us about effectiveness of binding & catalysis? (2) How can we determine experimentally the value of these constants for a given enzyme? II. Enzyme kinetics: Michaelis-Menten equation E+S k1 ES k2 E+P k-1 d[P] k2 [E]t [S] Initial reaction rate = ––– = vo = ––––––––– dt Km + [S] Michaelis-Menten equation [E]t = concentration of total enzyme [S] = concentration of free substrate Information obtained from the study of vo vs [S] k2: catalytic power of the enzyme (turnover rate), aka kcat; unit: 1/s Km: effectiveness (affinity) with which enzyme E binds S; unit: M Rate of breakdown of ES k-1 + k2 Km = –––––– k1 Rate of formation of ES III. How do we measure k2 and Km values? A. Typical experiment urease (0.1 M) (urea) + H2O CO2 + 2 NH3 Raw data urea (mM) 5 10 20 50 etc velocity/rate (M CO2/min) 30 50 80 100 Vmax 100 vo 50 [urea] III. Why is there a Vmax? urease (0.1 M) (urea) + H2O CO2 + 2 NH3 Vmax 100 vo 50 [urea] B. How do we get k2 and Km from this graph? Vmax vo k2 [E]t [S] vo = ––––––––– Km + [S] Vmax/2 Km [S] Consider three special cases 1. [S] = 0 vo = 0 2. [S] ≈ ∞ vo ≈ k2 [E]t = Vmax, so k2 = Vmax / [E]t Remember a finite number (Km) becomes negligible in the face of infinity 3. [S] = Km when vo = ½ Vmax Assumptions for steady-state kinetics E+S k1 k-1 ES k2 E+P The Michaelis-Menten equation assumes that the chemical reaction has reached steady state: • [ES] remains constant over time • presteady state (the build up of the ES complex) happens in microseconds • Usually nM [enzyme] but mM [substrate] in reaction, so [S] >> [E] k2 [E]t [S] vo = ––––––––– Km + [S] Vmax [S] with k2 [E]t = Vmax, then vo = ––––––––– Case 2 from previous slide! Km + [S] IV. What is the physical meaning of k2? Suppose [E]t = 0.01 µM, Vmax = 200 µM/min Vmax 200 µM/min k2 = –––––– = –––––––––––– = 20,000 min-1 [E]t 0.01 µM so 20,000 moles of P produced per min per mole of E k2 is the # of reactions a single enzyme molecule can catalyze per unit time k2 = kcat = “catalytic constant” or “turnover number”, expressed in catalysis events per time. V. What is the physical meaning of Km? E+S k1 k-1 ES k2 E+P Rate of breakdown of ES k-1 + k2 k-1 Km = –––––– ≈ ––– = Kdiss k1 k1 Rate of formation of ES provided (k2 << k-1) Remember: k2 is rate-limiting thus rate is slower than k-1 and thus k2 numerically much smaller than k-1 1. Km is a measure of how tightly an enzyme binds its substrate. 2. It is the value of [S] at which half of the enzyme molecules have their active sites occupied with S, generating ES. 3. For a given enzyme each substrate has its own Km. 4. Lower Km values mean more effective binding. Consider Km = 10-3 vs. 10-6 M (high affinity vs low affinity, compare to P s for T 50 and R states of hemoglobin, lecture 7) VI. A better way to plot vo vs [S] data. vo vs [S] plot Vmax Lineweaver-Burk plot ? vo 1/vo 1/Vmax Km [S] Vmax [S] vo = ––––––––– Km + [S] -1/Km 1/[S] 1 Km 1 1 –– = –––– ––– + ––––– vo Vmax [S] Vmax Lineweaver-Burk eliminates uncertainty in estimating Vmax. The estimates of Vmax and Km are thus greatly improved. Vmax [S] vo = ––––––––– Km + [S] Take reciprocal of both sides of equation Km + [S] 1 = –––––––– vo Vmax [S] Expand Thus [S] K 1 m + = –––––––– vo Vmax [S] Vmax[S] 1 Km 1 1 –– = –––– ––– + ––––– vo Vmax [S] Vmax Lineweaver-Burk y = ax + b Lineweaver-Burk plot Vmax [S] vo = ––––––––– Km + [S] 1/vo 1 Km 1 1 –– = –––– ––– + ––––– vo Vmax [S] Vmax y = a x + 1/Vmax -1/Km 1/[S] b Solve for y at x=1/[S]=0: 1 1 y = –– = –––– = b vo Vmax Solve for x at y=1/v0=0: 1 1 Vmax b x = –– = - –––– –– = - –– [S] Vmax Km a VII. Enzyme efficiency Efficiency = kcat / Km (specificity constant) Combines an enzyme’s catalytic potential with its ability to bind substrate at low concentration. Example – which enzyme is more efficient? Enzyme Km Chymotrypsin kcat kcat/Km 0.015 M 0.14 s-1 9.3 0.0003 M 0.50 s-1 1,700 Ac-Phe-Gly Ac-Phe + Gly Pepsin Phe-Gly Phe +Gly VIII. Enzyme inhibition - what to know 1. Reversible vs. irreversible inhibition • What is the difference? 2. Competitive inhibition • Know how to recognize or draw the model. • Know how vo vs [S], and Lineweaver-Burk plots are affected by competitive inhibition. • What are and Ki? 3. Irreversible inhibition • What is it; how does it work; what is its use? • What are suicide inhibitors, how do they work? • Know one example. Classical competitive inhibition where I is the inhibitor K1 How do you measure competitive inhibition? K1 -1/Km Vmax [S] vo = ––––––––– Km + [S] where [I] = 1 + ––– KI [E][I] KI = –––––– [EI] Vmax remains unchanged, but apparent Km increases with increasing [I] Inactivation of chymotrypsin by diisopropylfluorophosphate, an irreversible or suicide inhibitor Chymotrypsin is a serine protease that cleaves a peptide at Phe/Tyr/Trp (C) leaving a COO- on Phe/Tyr/Trp Inhibitor (diisopropylfluorophosphate) R2 Aspirin acts as an acetylating agent where an acetyl group is covalently attached to a serine residue in the active site of the cyclooxygenase enzyme, rendering it inactive.