* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Testing the ABC floral-organ identity model
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
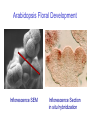
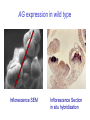
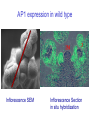
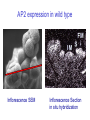
Testing the ABC floral-organ identity model: expression of A and C function genes Objectives: To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions concerning the phenotype of double or triple loss-offunction mutants and compare with the actual double mutant phenotypes. 2. Clone and sequence the ABC genes. Look for similarities with sequenced genes already in the database. 3. Determine the time and place of expression for each ABC gene and consider whether the expression correlates with the functional domain defined by the loss-of-function phenotype. 4. Test regulatory interactions between ABC genes by examining how the loss-of-function of one gene affects the expression domain of another. 5. Create gain-of-function mutants by generating transgenic plants carrying an ABC gene cDNA under the control of the CaMV35S promoter. Arabidopsis Floral Development Inflorescence SEM Mature Flower Flower Development Arabidopsis Floral Development Inflorescence SEM Inflorescence Section Arabidopsis Floral Development Inflorescence SEM Inflorescence Section in situ hybridization A Model For Control of Organ Type sepal petal 1 2 stamen carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) AG expression in wild type Inflorescence SEM Inflorescence Section in situ hybridization AG expression in wild type AG expression in wild type Mature Flower Inflorescence Section in situ hybridization A Model For Control of Organ Type sepal petal 1 2 stamen carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) AP1 expression in wild type Inflorescence SEM Inflorescence Section in situ hybridization AP1 expression in wild type AG expression AP1 expression A Model For Control of Organ Type sepal petal 1 2 stamen carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) AP2 expression in wild type Inflorescence SEM Inflorescence Section in situ hybridization AP2 expression in wild type A Model For Control of Organ Type Ap2 mutant carpel stamen stamen carpel 1 2 3 4 B (AP3, PI) C (AG) Where will AG be expressed? AG expression in an Ap2 mutant A Model For Control of Organ Type: Ag mutant sepal petal petal sepal 1 2 3 4 B (AP3, PI) A (AP1, AP2) Where will Ap1 be expressed? AP1 expression in an Ag mutant A Model For Control of Organ Type: Ag mutant sepal petal petal sepal 1 2 3 4 B (AP3, PI) A (AP1, AP2) Where will Ap2 be expressed? AP2 expression in an Ag mutant (unchanged) A Model For Control of Organ Type: 35SAG ? sepal ? petal 1 2 ? stamen ? carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) C (AG) Structure of wild type and mutant Arabidopsis flowers WT Whorl 1 Whorl 2 Whorl 3 Whorl 4 SEPAL PETAL STAMEN CARPEL ? STAMEN/ no organ STAMEN STAMEN CARPEL CARPEL 35S E ? Ag O SEPAL/ CARPEL 35S E AP2 O 35S E AP1 O A Model For Control of Organ Type: 35SAP2 or 35SAP1 ? sepal ? petal 1 2 ? stamen ? carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) A (AP1 OR AP2) Structure of wild type and mutant Arabidopsis flowers Whorl 1 Whorl 2 Whorl 3 Whorl 4 SEPAL PETAL STAMEN CARPEL 35S E ? AG O SEPAL/ CARPEL 35S E SEPAL AP2 O SEPAL ? STAMEN/ no organ PETAL PETAL STAMEN STAMEN CARPEL CARPEL STAMEN STAMEN CARPEL CARPEL 35S E SEPAL AP1 O SEPAL PETAL PETAL ? STAMEN ? CARPEL WT A Model For Control of Organ Type: 35SAP2 or 35SAP1 ? sepal ? petal 1 2 ? stamen ? carpel 4 3 B (AP3, PI) A (AP1, AP2) C (AG) A (AP1 OR AP2) Review of: Analysis of Class A and C expression. 1. In wild type plants, AP1 transcript is first observed throughout the stage 1 floral meristem but disappears from the centre of the stage 3 floral meristem. In later stages it continues to be expressed in developing sepals and petals. 2. In wild type plants, AP2 transcript is observed throughout the stage 1 floral meristem and continues to be found in all whorls in later stages of floral development. 3. In wild type plants, AG transcript is first observed in the central region of the stage 3 floral meristem. In later stages it continues to be expressed in developing stamens and carpels. 4. In Ap2 mutants, AG transcript appears in late stage 2/early stage 3 throughout the floral meristem although AG is most strongly expressed in the center of the floral meristem. 5. In Ag mutants AP1 transcript persists throughout the developing flower past stage three. The pattern of AP2 transcript in an Ag mutant is the same as in wild type. 6. Ectopic expression of AP2 or AP1 using the CaMV35S promoter does not affect floral organ type. Hypothesis for A-C function interactions 1. AP1 and AP2 appear early and are needed to specify perianth. 2. AG appears in stage 3 and negatively regulates expression of AP1 in inner cells and promotes formation of reproductive organs. 3. AP1 and AP2 prevent AG expression in perianth. 4. At least one other regulator must exist to promote AG expression in the centre at stage 3.