* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Open Access version via Utrecht University Repository
Immune system wikipedia , lookup
Major histocompatibility complex wikipedia , lookup
Antimicrobial peptides wikipedia , lookup
Monoclonal antibody wikipedia , lookup
DNA vaccination wikipedia , lookup
Adaptive immune system wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Innate immune system wikipedia , lookup
Adoptive cell transfer wikipedia , lookup
Molecular mimicry wikipedia , lookup
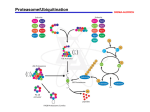
Lione Willems Master thesis October 2013 – December 2013 Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, The Netherlands Immunoproteasomes and their functions in immunity and cell viability Cover figure: Degradation of proteins into peptides by the proteasome Adapted from 1 Immunoproteasomes and their functions in immunity and cell viability Lione Willems Student number: 3384926 Utrecht University, Graduate School of Life Sciences, Infection & Immunity December 2013 Proteasomes are large protein complexes capable of degrading poly-ubiquitylated proteins. They function in the degradation of misfolded and damaged proteins and in many other cellular processes. Upon induction of the immunomodulatory cytokine interferon γ, the proteasomal catalytic subunits are substituted for the catalytic immunosubunits. These proteasomes are called immunoproteasomes and possess altered cleavage site specificity. For a long time they have only been implicated in antigen presentation. Immunoproteasomes were shown to be more efficient in the generation of antigenic peptides presented by MHC class I molecules. However, processing of several antigenic peptides required the presence of the standard proteasome and they were destroyed by immunoproteasomes, indicating that immunoproteasomes do not solely function in antigen presentation. Immunoproteasomes have indeed been implicated in other immune-related processes, including cytokine production. Interestingly, immunoproteasome expression has also been observed in non-immune cells and immune-privileged tissues, suggesting a function beyond immunity. Indeed, these proteasomes were found to be more efficient in the clearance of poly-ubiquitylated proteins and aggresomes upon stress and thereby in the maintenance of protein homeostasis and cell viability. Which of the immune or non-immune functions is executed by the immunoproteasomes might be dependent on the cell type expressing these proteasomes. Understanding the mechanisms of immunoproteasome functioning will be an important field of research and may help in the development of new treatments for many diseases. Supervisor: Dr. E.J.A.M. Sijts, Utrecht University, Faculty of Veterinary Medicine, Department of Infectious Diseases and Immunology Acknowledgements I want to express my thanks to my supervisor, Alice Sijts. She introduced me in the highly interesting subject of immunoproteasomes. I am thankful for her guidance and the feedback she gave me during the process of writing this thesis. Furthermore, I highly appreciate how fast she responded with feedback after handing in parts of the thesis. I would also like to thank dr. D.M.W. Zaiss for taking the time to review and asses this thesis. Contents Contents ............................................................................................................................................ 3 Abbreviations..................................................................................................................................... 4 Introduction ....................................................................................................................................... 5 Functions of immunoproteasomes in immune responses .................................................................... 7 Antigen presentation..................................................................................................................................7 Positive selection of CD8+ T cells in the thymus ........................................................................................9 Cytokine production ...................................................................................................................................9 Non-immune related function of immunoproteasomes: maintenance of protein homeostasis ........... 11 Significance of the immune and non-immune related functions of immunoproteasomes ................... 14 Treatment of pathologies .........................................................................................................................15 Concluding remarks ..................................................................................................................................15 References ....................................................................................................................................... 17 Abstract for laymen .......................................................................................................................... 20 Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 3 Abbreviations 26S proteasome ALIS cTEC DRiPs IFNγ LPS MHC TCR Standard proteasome Aggresome-like induced structure Cortical thymic epithelial cells Defective ribosomal products Interferon γ Lipopolysaccharide Major histocompatibility complex T cell receptor Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 4 Introduction The degradation of proteins is a major mechanism in the cell. Misfolded and damaged proteins are cleared to keep cells viable and a wide variety of cellular processes is regulated via protein degradation, including the cell cycle, apoptosis, transcription, metabolism and immune responses 2,3. It has been estimated that 80 to 90% of protein degradation in mammalian cells is achieved by a large protease complex called proteasome 2,4. Consequently, alterations in the activity and specificity of proteasomes have a great impact on the majority of cellular processes and may lead to diseases. The proteasome is part of the ubiquitin-proteasome system and is capable of degrading proteins in a fast and selective fashion 5. This is achieved by the ubiquitylation of protein substrates and subsequent recognition and degradation of the poly-ubiquitylated proteins by the proteasome (Figure 1). First, ubiquitin is activated and bound to a ubiquitin-activating enzyme, E1. A ubiquitin-conjugating enzyme, E2, is then responsible for transfer of the activated ubiquitin moiety to one of the E3 enzymes. These ubiquitin-protein ligases interact with protein substrates and catalyse the covalent binding of the ubiquitin moiety to this protein. By repeating these steps, additional ubiquitin molecules are attached to the previous ones and a poly-ubiquitin chain is formed. Finally, the poly-ubiquitylated proteins are recognized and degraded by the proteasome 6. Figure 1: Structure of 26S proteasome and substrate degradation (a) Schematic representation of 26S proteasome structure, containing the 20S core complex and a 19S regulatory complex. The 20S complex is composed of two rings of seven distinct α-subunits and two rings of seven distinct β-subunits. Proteasomal catalytic activity resides within the β-rings. In immunoproteasomes, the subunits β1, β2 and β5 are substitutes for the immunosubunits β1i, β2i and β5i respectively. (b) Schematic view of substrate degradation by the 26S proteasome. A poly-ubiquitylated protein is recognized by the 19S regulatory complex, followed by entry of the unfolded protein into the 20S core complex. The protein is cleaved into small peptides, which exit the proteasome on the other end. Adapted from 5. The type of proteasome most frequently used in the ubiquitin-proteasome system is referred to as the 26S proteasome and consists of a 20S core complex and a 19S regulatory complex. In eukaryotes, the core complex is a barrel-shaped structure built of four rings formed by two copies each of 14 different subunits. Seven distinct α-subunits (α1-7) associate into two identical α-rings located at the edges of the complex. The two identical inner rings are composed of seven distinct β-subunits (β1-7) (Figure 1a) 7. Assembly of the 20S core complex starts with the formation of the α-rings mediated by Proteasome Assembling Chaperones: PAC1-PAC2 dimers and PAC3. The β-rings are then assembled on top of the Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 5 α-rings by the addition of β-subunits in a specific order: β2, β3, β4, β5, β6, β1, β7. The resulting intermediates are composed of a single α-ring and β-ring. Five of the β-subunits (β1, β2, β5, β6 and β7) in these intermediates express a propeptide that needs to be processed to complete proteasomal maturation. Maturation is accomplished by dimerization of the intermediates which induces autocatalytic cleavage of the β-subunit propeptides and finishes the 20S core complex assembly 2,3. Proteolytic activity of the 20S proteasome resides within the β1, β2 and β5 subunits and cleavage of their propeptides exposes the catalytic Threonine1 residues 7. These three β-subunits differ in their catalytic activity. β1 is associated with proteolysis immediately after the acidic amino acid aspartic acid, which is described as caspase-like activity (also sometimes referred to as PGPH activity). Subunit β2 has been associated with trypsin-like activity and results in cleavage after the basic amino acids lysine and arginine. At last, the catalytic site of β5 has been shown to cleave after hydrophobic or aromatic amino acids (tyrosine, tryptophan and phenylalanine) and is named chymotrypsin-like activity 7,8. Proteolytic cleavage occurs at the inner surface of the 20S core complex and therefore protein substrates need to enter this complex via the opening in the α-rings. However, entry via this opening is blocked by N-terminal protrusions of the subunits α1, α2, α3, α6 and α7. It is thought that access requires rearrangements triggered by the 19S regulatory complexes 7. This complex associates with the α-subunits of the core complex and is composed of ATPase and non-ATPase subunits. One of the ATPases (Rpt5) binds poly-ubiquitylated protein substrates, followed by ATP-dependent de-ubiquitylation. The remaining substrates are still too big though to enter the 20S core complex and ATP-dependent unfolding enables them to fit within the channel. At last, entry is induced by opening the channel which requires ATP hydrolysis 2,3,5. Specificity of the 26S proteasome is achieved by selective ubiquitylation of protein substrates and recognition of the poly-ubiquitin chain by the regulatory 19S complexes. Especially the E3 enzymes seem to play a major role in specificity. A large group of these enzymes exist in mammals and each of them recognizes a variety of protein substrates. Expression of distinct E3 enzymes at different intracellular locations results in poly-ubiquitylation and proteasomal cleavage of specific substrates. Specificity is further enhanced by post-translational modification and associating proteins which may be important in the recognition of protein substrates by E3 enzymes 5,6. Specificity of the proteasome can also be altered by the expression of three additional catalytic β-subunits (β1i, β2i and β5i), termed immunosubunits. Their expression is induced by the immunomodulatory cytokine interferon γ (IFNγ) and they are incorporated into the 20S core complex, thereby replacing the standard β1, β2 and β5 catalytic subunits. The resulting proteasome is referred to as the immunoproteasome 9. Assembly of the immunoproteasome is slightly different from the standard (26S) proteasome. It starts with β1i, which facilitates incorporation of β2i. Next, β3 and β4 are incorporated, followed by β5i. The propeptide of β5i is required for cleavage of the β1i and β2i propeptides and is essential in immunoproteasome maturation. At last, β6 and β7 are incorporated 3,10,11. The immunoproteasome enhances the generation of antigenic peptides presented by MHC class I molecules if compared with the standard proteasome 12. In agreement with this function, immunoproteasomes are constitutively expressed in several types of immune cells, including the professional antigen presenting cells macrophages and dendritic cells. However, constitutive expression has also been observed in non-immune tissues, including the liver and colon 13. These observations suggest immunoproteasomes are also involved in processes different from antigen presentation. Here, we will review the role of immunoproteasomes in immunity and its non-immunological role in protein homeostasis. Furthermore, the significance of its function in protein homeostasis compared with the immunological functions will be discussed. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 6 Functions of immunoproteasomes in immune responses Antigen presentation Human cells present antigenic peptides on its surface via major histocompatibility complex (MHC) class I molecules. These peptides are generated by the degradation of intracellular proteins by proteasomes in the cytoplasm and subsequent translocation to the endoplasmic reticulum and binding to MHC class I molecules (Figure 2). In case of viral infections, viral proteins are degraded and via this process of antigen presentation expressed on the cell’s surface. The MHC molecules bound to these viral peptides are then recognized by antigen-specific CD8+ T cells and elicit an immune response in order to clear the infected cells. Several cell types, including macrophages and dendritic cells, are known to be specialized in antigen presentation. Besides presentation of intracellular proteins, they are capable of presenting exogenous proteins on MHC class II molecules upon endocytosis and degradation. Thereby, immune responses are regulated. The first indication that immunoproteasomes play a role in antigen presentation, was the finding that immunosubunits β1i and β5i are located within the human MHC class II region 14,15. Indeed, analysis of the generation of a hepatitis B virus epitope has revealed that its production requires the presence of the IFNγ-inducible immunoproteasome 16 and also production of the ovalbumin epitope was shown to be more efficient by immunoproteasomes 17. Moreover, deletion of immunosubunits has been reported to result in reduced cell surface expression of MHC class I molecules and in altered efficiency in the presentation of peptides 18,19. Abundance and diversity of antigenic peptides was increased in immunoproteasome-expressing cells, and may contribute to a higher efficiency of antigen presentation observed in presence of immunoproteasomes 20. It has been suggested that immunoproteasomes possess two features resulting in increased abundance and diversity of the peptides: altered cleavage site specificity and enhanced rate of proteasome-mediated digestion if compared with standard proteasomes 16,20. Recently published results on the comparison of the standard and immunoproteasome crystal structures confirmed these suggestions 9. Substitution of β1 for the β1i subunit increases the hydrophobic character of the proteasome and as a consequence β1i favours cleavage after small, hydrophobic residues and the proteasome (partially) loses its caspase-like activity. Comparison of β5 and β5i subunits showed that residue 27 is substituted for a more hydrophilic residue in β5i. In combination with other amino acid substitutions this results in a chymotrypsin-like activity for β5i, while β5 resembles a more elastase-like activity. For β2i its structure and catalytic function is similar to that of the standard β2 subunit and the relevance for its incorporation is unclear. Nonetheless, trypsin-like activity is increased in immunoproteasomes and might be the result of conformational changes. Overall, immunoproteasomes possess lower caspase-like activity and higher chymotrypsin- and trypsin-like activities. Further evidence for the impact of immunoproteasomes on antigen presentation was found by analysis of triple knockout mice, lacking all three immunosubunits 21. Out of 11 antigenic peptides validated, 8 peptides were observed to be lower expressed by MHC class I molecules in absence of the immunosubunits. It seems that certain peptides are efficiently generated by the immunoproteasome, while they are not in presence of the standard proteasome only. As a result, different sets of antigenic peptides are generated by these two types of proteasomes. It should be noted though that while many peptides are efficiently produced by the immunoproteasome, some are preferentially generated by the standard proteasome and their generation by the immunoproteasome is highly inefficient 22. For example, gp100 and tyrosinase antigenic peptides are produced by the standard proteasome, but not the immunoproteasome, and cells exclusively expressing these standard proteasomes efficiently stimulate gp100- and tyrosinase-specific cytotoxic T lymphocytes respectively. Absence of the antigenic peptides in immunoproteasome-expressing cells was found to be the result of destructive cleavages behind internal hydrophobic residues, which is in agreement with the higher chymotrypsin-like activity of Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 7 the immunoproteasome. In contrast, MAGE-C2 antigenic peptide is preferentially generated by the immunoproteasome. The higher caspase-like activity of standard proteasomes results in destructive internal cleavage of the peptide after an acidic residue. Taken together, the changed subunit composition of immunoproteasomes relative to standard proteasomes seems to contribute to altered protease activities and differential processing of antigenic peptides, resulting in the generation of different antigenic peptides by these two types of proteasomes. Figure 2: The role of the (immuno)proteasome in antigen presentation Poly-ubiquitylated cytoplasmic proteins are recognized and degraded by either the standard, the immuno- or the intermediate proteasome. Each of the proteasome-types generates another set of antigenic peptides. The peptides are transported into the endoplasmic reticulum by a complex called TAP and attached to MHC class I molecules. Peptide-MHC complexes are then transported to the cell surface where specific CD8+ T cells may recognize one of the presented peptides and elicit an immune response. Adapted from 1. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 8 Diversity of the antigenic peptides might be further increased by the expression of intermediate proteasomes 23,24. They contain a combination of standard and immunosubunits and, as can be expected, show slightly different activities 23. Mainly the caspase-like activity is changed, dependent on the incorporation of β1i or not. Due to the altered activities, a different set of peptides suitable for antigen presentation is generated. While some peptides are preferentially produced by one of the intermediate proteasomes, others are internally cleaved and require their absence 23,25,26. The presence of the different types of proteasomes might be important in shaping the CD8+ T cell responses, by increasing the diversity of antigenic peptides presented on MHC class I molecules and inducing cell type-specific expression of antigenic epitopes 26. Positive selection of CD8+ T cells in the thymus Presentation of foreign antigenic peptides via MHC class I molecules elicits CD8+ T cell responses and clearance of the cells presenting these foreign peptides. As described above, proteasomes are involved in the generation of the peptides presented on the cell surface, but recently they have also been implicated in the development of the CD8+ T cells in the thymus 27,28. T cell development starts with the positive selection of double-positive thymocytes (CD4+CD8+) in the thymic cortex, resulting in CD4 or CD8 single-positive thymocytes. During this selection process, CD4+CD8+ thymocytes interact with cortical thymic epithelial cells (cTECs). cTECs express MHC molecules loaded with peptides. Weak interactions between these peptide-MHC complexes and the T cell receptor (TCR) present on thymocytes induce survival of the thymocytes and selection into either CD4 or CD8 single-positive cells. The next step in the development is negative selection during which single-positive thymocytes that strongly interact with peptide-MHC complexes are eliminated by the induction of apoptosis. Negative selection prevents the development of T cells recognizing self-peptides and the induction of autoimmune diseases. A specific type of proteasome, named thymoproteasomes, seems to be important in the positive selection of double-positive thymocytes. Thymoproteasomes contain the immunosubunits β1i and β2i and a catalytic subunit which is exclusively found in cTECs, β5t. As do the immunosubunits, the β5t subunit changes the cleavage specificity of the proteasome. The caspase-like and trypsin-like activities remain unchanged, however thymoproteasomes possess clearly reduced chymotrypsin-like activity if compared with standard proteasomes 27. This reduction in chymotrypsin-like activity might result in the generation of peptides that bind MHC class I molecules with low affinity, thereby forming unstable peptide-MHC complexes. As low-affinity interactions between TCR and peptide-MHC complexes drive positive selection of thymocytes, it has been suggested that these unstable complexes are important in this specific step of thymocyte development. Thymoproteasomes thus seem to play a role in the generation of peptides suitable for positive selection 28. Relevance of the β5t subunit and the thymoproteasome has been observed in mice deficient for β5t. Reduced numbers of CD8+ T cells were found in the thymus and periphery and high lethality was shown upon influenza virus infection 27–29. It can be suggested that β5t-deficiency results in inefficient selection and generation of a CD8+ T cell repertoire and consequently in susceptibility to infections. Cytokine production Contrary to what was initially suggested, immunoproteasomes were found to not solely function in antigen presentation. Immunosubunits were identified in thymoproteasomes which are important for the generation of a CD8+ T cell repertoire and recent evidence suggests yet another function in immunity for immunoproteasomes: promoting the production of cytokines 30,31. To elicit an immune response, murine macrophages were stimulated with lipopolysaccharide (LPS) which is a compound found in the outer membrane of gram-negative bacteria and induces the increased Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 9 expression of immunosubunits and immunoproteasomes. The production of several pro-inflammatory mediators (such as NO, iNOS, IRF3 and STAT1/3) and cytokines (such as IFNγ, IL-1β and IL-6) was induced in these macrophages, while reduced production was observed in macrophages isolated from β2i/β5i double knockout mice 32. Also selective inhibition of the β5i subunit, but not the β5 subunit, resulted in a decrease in the production of TNFα, IL-23 and IL-6 cytokines by LPS-stimulated peripheral bone marrow cells 33, indicating a potential function of the immunosubunits in the production of these pro-inflammatory mediators and cytokines. Confirming this observation, the same authors report reduced cytokine production by splenocytes from wild type mice but not β5i-deficient mice upon treatment with the β5i inhibitor. Interestingly, in the same experiment no decrease in cytokine production was observed in β5i-deficient murine splenocytes in absence of the selective inhibitor if compared with untreated splenocytes from wild type mice. As discussed by Sijts and Kloetzel 34, the standard β5 subunit is probably able to replace the non-functional β5i subunit in β5i-deficient mice and to adopt the function of β5i in cytokine production. The decrease in cytokine production observed in wild type splenocytes upon treatment with the β5i-specific inhibitor might be explained by the inability to replace the inhibited β5i subunits with the standard β5 subunits. Therefore, these findings suggest an important function of immuno- and standard proteasomes in the production of cytokines. The mechanisms of proteasome-induced cytokine production remain unclear though. It is possible that proteasomes cleave a factor which regulates cytokine production. Indeed, proteasomes were found to function in the activation of the transcription factor NFκB, which regulates the expression of cytokines amongst others 35. Recently, also immunoproteasomes have been implicated in NFκB activation 36. In the gut epithelium of patients with Crohn’s disease, which is associated with enhanced cytokine production, increased expression of immunoproteasomes has been correlated with enhanced processing of p105 and degradation of IκBα and subsequent activation of NFκB. Furthermore, co-localization of immunosubunit β2i and the active NFκB complex in the intestinal mucosa of these patients suggests immunosubunits regulate NFκB activation 36. In line with this, another report shows a reduction in cytokine levels and a decrease in the degradation of IκBα in β1i-deficient dendritic cells upon infection with influenza A virus 37. On the contrary, it has also been reported that selective β5i inhibition in a TNFα-stimulated reporter cell line did not reduce NFκB activation, suggesting NFκB-independent induction of cytokine production by the immunoproteasome 33. Possibly several pathways may be altered by the action of immunoproteasomes, resulting in the regulation of cytokine production. Increased levels of cytokines are found in patients with Crohn’s disease and ulcerative colitis. Selective inhibition of β5i has been shown to reduce cytokine levels, inflammation and tissue destruction in mice models of these two autoimmune diseases, indicating that immunoproteasomes promote the development of these autoimmune diseases by the induction of cytokine production 36,38. It might not be a direct effect of immunoproteasomes though. Kalim et al. 39 report that deficiency and selective inhibition of β5i in mice supresses the differentiation of the inflammatory Th17 cells. Th17 cells have been implicated in the development of autoimmune diseases and are known to induce the production of pro-inflammatory cytokines 40. The reduced levels of cytokines found in Crohn’s disease and ulcerative colitis upon β5i-inhibition might therefore be the result of the suppression of Th17 cell differentiation and not by direct inhibition of cytokine production. Simultaneously, β5i-deficiency or inhibition induces the differentiation of regulatory T cells 39. These cells have a suppressive effect on the proliferation of CD4+ and CD8+ T cells and thereby reduce immune responses directed against self-antigens. All together these data indicate that functional immunoproteasomes may promote the development of autoimmune diseases by indirectly inducing the production of pro-inflammatory cytokines and by removing the suppressive effect of regulatory T cells. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 10 Non-immune related function of immunoproteasomes: maintenance of protein homeostasis Conforming their function in immune responses, tissue expression of immunoproteasomes was found to be the highest in the spleen, which is involved in the initiation of immune responses upon recognition of pathogens in the blood. Furthermore, immunoproteasomes were found to be constitutively expressed in several hematopoietic cells, including macrophages, B cells and dendritic cells 13. Though, immunosubunits were also abundantly observed in non-immune cells and tissues. Their expression in tissues like colon, liver and kidneys and more interestingly in immune-privileged tissues like brain and eyes suggests immunoproteasome functions distinct from their functions in immunity 13,41,42. Recently published results have implied their significance in cell viability by the removal of damaged and aggregated proteins 43. Within the cell damaged, misfolded or prematurely terminated proteins become poly-ubiquitylated and form a pool of defective ribosomal products (DRiPs). Under normal conditions, these defective proteins are degraded by the standard proteasome. However, stress conditions, e.g. infections, starvation, oxidative stress and ageing, induce excessive production of DRiPs and the standard proteasomes do not have the capacity to clear this increased amount of DRiPs 43,44. As a consequence, the poly-ubiquitylated proteins form aggresome-like induced structures (ALIS), which can be harmful for the cell by disrupting cellular processes 45. Seifert et al. 43 reported that interferon-induced oxidative stress in murine fibroblasts and human HeLa cells resulted in an initial increase of the levels of poly-ubiquitylated peptides, but was followed by a decline after 24 hours and a return to the levels before stress induction. The decrease observed after 24 hours coincided with an increase in proteasomal chymotrypsin-like activity and the induction of immunoproteasomes, suggesting that immunoproteasomes function in the degradation of the poly-ubiquitylated peptides. Indeed, they were found to be at least 2-fold more efficient in substrate degradation and upon knockout of immunosubunit β5i, peptides accumulated and ALISs were formed. To examine the consequence of peptide accumulation and ALIS formation in the β5i-deficient mice, caspase activity was measured as an indication for the induction of apoptosis and was found to be increased upon stress. The authors hypothesized that oxidative stress induced by interferons results in the oxidation and poly-ubiquitylation of proteins and the formation of a large pool of DRiPs. In the absence of functional immunoproteasomes this would cause the formation of toxic ALISs and subsequently the induction of apoptosis. However, interferons additionally induce the formation of immunoproteasomes. These proteasomes efficiently degrade poly-ubiquitylated peptides and thereby prevent the formation of ALISs and the induction of apoptosis (Figure 3) 43. Immunoproteasomes thus seem to control protein homeostasis upon stress. The results described in above mentioned report were questioned by Nathan et al. 46. They argued that IFNγ stimulation does not result in an increase in the levels of poly-ubiquitylated peptides and in the formation of ALISs as they were not able to detect either of the two. They also failed to confirm the data on increased degradation efficiency of immunoproteasomes. Based on these results, the authors concluded that immunoproteasomes are not more important in protein homeostasis than standard proteasomes. Discrepancies between both reports, however, might be explained by distinct experimental procedures 47. Furthermore, similar results as reported by Seifert et al. confirming the significance of immunoproteasomes in protein homeostasis upon stress induction have been published 41,42,48,49 . Increased synthesis of immunoproteasomes was observed following oxidative stress in murine fibroblasts 48 and in bovine and human aortic endothelial cells 49. In the latter, this upregulation was shown to be dependent on signaling via the second messengers cAMP and cGMP and the protein kinases PKA and PKG, and resulted in the inhibition of oxidative stress and apoptosis. By maintaining protein Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 11 homeostasis, immunoproteasomes thus prevent the induction of apoptosis and have a significant function in the cell’s viability. Interestingly, immunoproteasome expression and its significance in cell viability was also observed in immune-privileged tissues like the brain and the retina in the eyes 41,42. In these tissues, an impaired response to oxidative stress and an increased susceptibility to cell death emerged in response to immunosubunit deficiency. These reports further support the idea of non-immune related functions of immunoproteasomes. Figure 3: The role of the (immuno)proteasome in protein homeostasis In order to be functional, newly synthesized proteins need to be correctly folded. Misfolded and defective proteins (DRiPs) will be marked with a poly-ubiquitin chain and cleared by proteasomes. In absence of stress signals, like infections, oxidative stress and ageing, the standard proteasome fulfills this task of protein homeostasis (left). However, in presence of these stress signals, translation is enhanced and a larger number of newly synthesized proteins is oxidized. Standard proteasomes cannot keep up with this excessive production of DRiPs, resulting in the formation of the toxic aggresome-like induced structures (ALISs). Nevertheless, during stress the cytokine IFNγ is secreted which induces the formation of immunoproteasomes. The immunoproteasomes are capable of efficiently degrading the poly-ubiquitylated DRiPs. Thereby, they prevent the formation of ALISs and protect the cell against the toxic effects of these structures (right). Adapted from 43. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 12 A condition which is thought to be related to stress is ageing. In brain and liver of mice, an increase of oxidized proteins was observed with age, despite of a concomitant upregulation of proteasomal subunits, including the immunosubunits. Nonetheless, a significantly higher increase in protein oxidation was seen in β1i-deficient mice of the same age, indicating that immunoproteasomes do protect the cells against protein oxidation during ageing 41. Studies in the long-lived naked mole rat seem to confirm this finding 50. These rodents have an exceptionally long lifespan of about 32 years and maintain a good health for most of these years. Compared with mice, increased chymotrypsin-like and trypsin-like activities were found in liver fractions of the naked mole rats 50, which is a feature of immunoproteasomes 9. Indeed, immunoproteasome transcription was upregulated and might be responsible for protection against age-related protein oxidation and maintenance of health over many years in these naked mole rats 50. It might be questioned though whether it is reliable to compare naked mole rats with mice, even though they were matched according physiological age. In humans, impaired immunoproteasome functioning has also been linked to several pathologies. Nakajo-Nishimura syndrome 51 and Japanese autoinflammatory syndrome with lipodystrophy 52 are both caused by mutations in the β5i subunit. Due to the mutations, immunoproteasome assembly is disturbed and the chymotrypsin-like activity reduced. Consequently, oxidized and ubiquitylated proteins are not efficiently degraded in these patients. They accumulate and ALISs are formed, which may then induce the inflammatory conditions. Correct functioning of immunoproteasomes thus seems to be important in the maintenance of protein homeostasis and in the prevention of uncontrolled inflammation. This protective function of immunoproteasomes has also been observed upon viral infections. Coxsackievirus B3 can infect heart muscle cells and cause myocarditis. In immunosubunit β5i-deficient mice, more severe injury was observed upon infection. Oxidized and ubiquitylated proteins accumulated in these mice and NFκB activation was found to be impaired. Furthermore, the β5i-deficient heart cells were more sensitive to apoptosis 53. Together, these results strongly support a role for immunoproteasomes in protein homeostasis and cell viability. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 13 Significance of the immune and non-immune related functions of immunoproteasomes Substitution of the catalytic subunits β1, β2 and β5 for the respective immunosubunits results in altered cleavage site specificity and an enhanced rate of proteasome-mediated digestion. These immunoproteasomes possess lower caspase-like activity and higher chymotrypsin-like and trypsin-like activities and as a consequence generate a distinct set of cleaved peptides compared with standard proteasomes 9,16,20. The peptides generated by immunoproteasomes are more suitable for presentation on MHC class I molecules, while standard proteasomes are mainly required for maintaining protein homeostasis and regulating cellular processes. However, it was shown later that immunoproteasomes induce internal destructive cleavages in some peptides and presentation of these peptides is more efficient in presence of standard proteasomes 22. Expression of either standard or immunoproteasomes might be a way of regulating which peptides are presented at the cell surface and thus by which CD8+ T lymphocytes these cells can be recognized. Further fine-tuning of these T cell responses is achieved by the expression of intermediate proteasomes. Due to their altered cleavage site specificities, generation of several antigenic peptides is promoted in presence of one of these proteasomes, while for other peptides their absence is required. It thus seems that immunoproteasomes are not the only ones responsible for the presentation of peptide-MHC complexes. The expression of various proteasome types seems to result in an increased diversity of the antigenic peptides presented by distinct cells, which might help in inducing the appropriate immune responses 26. It should be studied though whether intermediate proteasomes are actually expressed by professional antigen presenting cells and indeed diversify CD8+ T cell responses. Yet another set of antigenic peptides is generated by the thymoproteasomes. Expression of the immunosubunits β1i and β2i, in combination with a subunit exclusively expressed in the thymus (β5t), results in reduced chymotrypsin-like activity compared with standard proteasomes. Due to this reduced activity, thymoproteasome-generated peptides are thought to have low affinity for MHC class I molecules and to form unstable complexes with MHC. These unstable complexes seem to be relevant in positive selection of CD8+ T cells in the thymus, which suggests that thymoproteasomes are important in shaping the T cell repertoire and immune responses 27,28. The immune-related functions of immunoproteasomes are further expanded by recent reports showing their involvement in the production of pro-inflammatory cytokines. Immunoproteasomes have been suggested to function in the activation of the transcription factor NFκB, which regulates the expression of a variety of immune-related genes including cytokines 36,37. Though, NFκB-independent induction of pro-inflammatory cytokines is also likely to occur 33. Possibly, several pathways are involved in the regulation of cytokine production. Which pathways are activated might be dependent on the expression of cell type-specific proteins and environmental factors and signals. Also the expression of intermediate proteasomes might result in the induction of different pathways of cytokine production. However, this has not been studied yet. Immunoproteasomes have been linked to immunological functions for a long time, but recently also a non-immunological role has emerged. They were suggested to play an important role in protein homeostasis upon stress conditions 43. During conditions like infections, oxidative stress and ageing, proteins are oxidized and may form aggregates, which are toxic for the cell. To maintain the cell’s viability, immunoproteasomes are induced. They remove the oxidized and aggregated proteins and thereby regulate protein homeostasis. Regulation of protein homeostasis by these proteasomes is observed in many cell types, including immune-privileged cells 41,42, and indicates a significant function of immunoproteasomes beyond immune responses. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 14 Treatment of pathologies Immunoproteasomes seem to play a role in the development of several pathologies. Conflicting data have been published though. These proteasomes were shown to promote the development of autoimmune diseases like Crohn’s disease and ulcerative colitis by the induction of cytokine production. In these patients, treatment with immunoproteasome inhibitors is suggested to relieve the symptoms and reduce the tissue destruction 36,38. On the contrary, immunoproteasomes seem to be protective in the development of autoimmune diseases in certain conditions 54. β5i and β2i double gene-deficient mice develop CD8+ T cell-mediated autoimmune diseases (including diabetes insipidus and diabetes mellitus) after irradiation and bone marrow transplantation. As discussed by Zaiss et al. 54, their results suggest that the higher chymotrypsin-like activity of functional immunoproteasomes would result in the generation of antigenic peptides capable of binding MHC class I molecules with higher affinity. As a consequence, formation of low-affinity interactions between self-recognizing CD8+ T cells and these peptide-MHC complexes is prevented and the chance of developing an autoimmune disease is reduced. In this case, patients would benefit from increased induction of functional immunoproteasomes. A protective function of immunoproteasomes has also been suggested in several other pathologies, e.g. Nakajo-Nishimura syndrome 51 and heart failure upon Coxsackievirus B3 infection 53, and in ageing 41,50. In these patients and in the elderly, immunoproteasomes were shown to be responsible for maintaining protein homeostasis and preventing apoptosis of the cells. Again, increased induction of functional immunoproteasomes might be helpful. Concluding remarks The desired treatment, either inhibition or induction of functional immunoproteasomes, seems to be dependent on which specific cellular process is involved in the development of the disease. Whether immunoproteasomes induce either excessive production of cytokines, efficient generation of antigenic peptides or maintenance of protein homeostasis, might be dependent on the cell type due to the presence of cell type-specific proteins and regulators and regulation of their expression by environmental factors and signals. Constitutive expression in professional antigen presenting cells might be relevant for antigen presentation and cytokine production. Expression in non-immune cells and, more interestingly, in immune privileged cells suggests a role in the viability of these cells by maintaining protein homeostasis upon stress. Moreover, fine-tuning of these processes might occur upon expression of intermediate proteasomes, as the altered cleavage site specificities possessed by the various proteasome types are likely to result in variations in the regulation of these processes (Figure 4). Though, more studies will have to be conducted to understand the exact mechanisms of regulation by intermediate and immunoproteasomes. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 15 Figure 4: Immune and non-immune related functions of (immuno)proteasomes A pool of poly-ubiquitylated proteins, composed of DRiPs, antigens, cell cycle regulators amongst others, exists in the cell. Antigens are degraded by either standard proteasomes or immunoproteasomes, resulting in the generation of MHC class I antigenic peptides. For immunoproteasomes also other functions have been implied. Their expression in hematopoietic cell types might modulate the expression of cytokines, possibly by regulating the activation of transcription factor NFκB. However, NFκB-independent pathways have also been suggested. The expression of immunoproteasomes is also likely to result in maintenance of protein homeostasis and cell viability. This function was suggested to be important for immune cells, but also non-immune cells including immune-privileged cells. Adapted from 13. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 16 References 1. Yewdell, J. W., Reits, E. & Neefjes, J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 (2003). 2. Voges, D., Zwickl, P. & Baumeister, W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 (1999). 3. Murata, S., Yashiroda, H. & Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 (2009). 4. Craiu, a. Lactacystin and clasto-Lactacystin beta -Lactone Modify Multiple Proteasome beta -Subunits and Inhibit Intracellular Protein Degradation and Major Histocompatibility Complex Class I Antigen Presentation. J. Biol. Chem. 272, 13437–13445 (1997). 5. Pickart, C. M. & Cohen, R. E. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 (2004). 6. Ciechanover, A. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO 17, 7151–7160 (1998). 7. Groll, M. et al. Structure of 20s proteasome from yeast at 2.4 Å resolution.pdf. Nature 386, 463–471 (1997). 8. Heinemeyer, W. The Active Sites of the Eukaryotic 20 S Proteasome and Their Involvement in Subunit Precursor Processing. J. Biol. Chem. 272, 25200–25209 (1997). 9. Huber, E. M. et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 148, 727–738 (2012). 10. Griffin, B. T. A. et al. Immunoproteasome Assembly : Cooperative Incorporation of Interferon gamma inducible subunits. J. Exp. Med. 187, 97–104 (1998). 11. Kingsbury, D. J., Griffin, T. a & Colbert, R. a. Novel propeptide function in 20S proteasome assembly influences beta subunit composition. J. Biol. Chem. 275, 24156–24162 (2000). 12. Kloetzel, P. M. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat. Immunol. 5, 661–669 (2004). 13. Ebstein, F., Kloetzel, P.-M., Krüger, E. & Seifert, U. Emerging roles of immunoproteasomes beyond MHC class I antigen processing. Cell. Mol. Life Sci. 69, 2543–2558 (2012). 14. Glynne, R. et al. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature 353, 357–360 (1991). 15. Kelly, A. et al. Second proteasome-related gene in the human MHC class II region. Nature 353, 667–668 (1991). 16. Sijts, a J. et al. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J. Exp. Med. 191, 503–514 (2000). 17. Cascio, P., Hilton, C., Kisselev, a F., Rock, K. L. & Goldberg, a L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 20, 2357–2366 (2001). 18. Rock, K. L. & Goldberg, a L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17, 739–79 (1999). 19. Groettrup, M. et al. The Interferon-gamma-inducible 11 S Regulator (PA28) and the LMP2/LMP7 Subunits Govern the Peptide Production by the 20 S Proteasome in Vitro. J. Biol. Chem. 270, 23808–23815 (1995). 20. De Verteuil, D. et al. Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol. Cell. Proteomics 9, 2034–2047 (2010). Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 17 21. Kincaid, E. Z. et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 13, 129–135 (2012). 22. Chapiro, J. et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J. Immunol. 176, 1053–1061 (2006). 23. Guillaume, B. et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. U. S. A. 107, 18599–18604 (2010). 24. De, M. et al. Beta 2 subunit propeptides influence cooperative proteasome assembly. J. Biol. Chem. 278, 6153–6159 (2003). 25. Guillaume, B. et al. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J. Immunol. 189, 3538–3547 (2012). 26. Zanker, D., Waithman, J., Yewdell, J. W. & Chen, W. Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. J. Immunol. 191, 52–59 (2013). 27. Murata, S. et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349– 1353 (2007). 28. Xing, Y., Jameson, S. C. & Hogquist, K. a. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc. Natl. Acad. Sci. U. S. A. 110, 6979–6984 (2013). 29. Nitta, T. et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32, 29–40 (2010). 30. Qureshi, N., Morrison, D. C. & Reis, J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta 1823, 2087–2093 (2012). 31. Basler, M., Kirk, C. J. & Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 25, 74–80 (2013). 32. Reis, J. et al. The immunoproteasomes regulate LPS-induced TRIF/TRAM signaling pathway in murine macrophages. Cell Biochem. Biophys. 60, 119–26 (2011). 33. Muchamuel, T. et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 15, 781–787 (2009). 34. Sijts, E. J. a M. & Kloetzel, P. M. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. C. 68, 1491–1502 (2011). 35. Palombella, V. J., Rando, O. J., Goldberg, A. L. & Maniatis, T. The Ubiquitin-Proteasome Pathway Is Required for Processing the NF-KB1 Precursor Protein and the Activation of NF-KB. Cell 78, 773–785 (1994). 36. Visekruna, A. et al. Proteasome-mediated degradation of I κ B α and processing of p105 in Crohn disease and ulcerative colitis. J. Clin. Invest. 116, 3195–3203 (2006). 37. Hensley, S. E. et al. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J. Immunol. 184, 4115–4122 (2010). 38. Basler, M., Dajee, M., Moll, C., Groettrup, M. & Kirk, C. J. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J. Immunol. 185, 634–641 (2010). 39. Kalim, K. W., Basler, M., Kirk, C. J. & Groettrup, M. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. J. Immunol. 189, 4182– 4193 (2012). 40. Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V. K. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517 (2009). Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 18 41. Ding, Q., Martin, S., Dimayuga, E., Bruce-keller, A. J. & Keller, J. N. LMP2 Knock-Out Mice Have Reduced Proteasome Activities and Increased Levels of Oxidatively Damaged Proteins. Antioxid. Redox Signal. 8, 130– 135 (2006). 42. Hussong, S. a, Kapphahn, R. J., Phillips, S. L., Maldonado, M. & Ferrington, D. a. Immunoproteasome deficiency alters retinal proteasome’s response to stress. J. Neurochem. 113, 1481–1490 (2010). 43. Seifert, U. et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142, 613–624 (2010). 44. Aiken, C. T., Kaake, R. M., Wang, X. & Huang, L. Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteomics 10, R110.006924 (2011). 45. Hartl, F. U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). 46. Nathan, J. a et al. Immuno- and constitutive proteasomes do not differ in their abilities to degrade ubiquitinated proteins. Cell 152, 1184–1194 (2013). 47. Ebstein, F. et al. Immunoproteasomes are important for proteostasis in immune responses. Cell 152, 935– 937 (2013). 48. Pickering, A. M. et al. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 432, 585–594 (2010). 49. Kotamraju, S. et al. Upregulation of immunoproteasomes by nitric oxide: potential antioxidative mechanism in endothelial cells. Free Radic. Biol. Med. 40, 1034–1044 (2006). 50. Rodriguez, K. a, Edrey, Y. H., Osmulski, P., Gaczynska, M. & Buffenstein, R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS One 7, e35890 (2012). 51. Arima, K. et al. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. U. S. A. 108, 14914–14919 (2011). 52. Kitamura, A. et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Invest. 121, 4150–4160 (2011). 53. Opitz, E. et al. Impairment of immunoproteasome function by β5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 7, e1002233 (2011). 54. Zaiss, D. M. W., Bekker, C. P. J., Gröne, A., Lie, B. a & Sijts, A. J. a M. Proteasome Immunosubunits Protect against the Development of CD8 T Cell-Mediated Autoimmune Diseases. J. Immunol. Balt. Md 1950 187, 2302–2309 (2011). Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 19 Abstract for laymen Proteasomes are large protein complexes capable of degrading marked proteins. A chain of several ubiquitin moieties serves as the mark of degradation. This poly-ubiquitin chain is recognized by the proteasome, which then cleaves the poly-ubiquitylated protein into small peptides. Proteasome-mediated degradation is important in the removal of misfolded and damaged proteins and in many other cellular processes, like immunity. Three proteasomal subunits possess cleavage activity and upon expression of the immunomodulatory cytokine interferon γ these three subunits are replaced by three immunosubunits, thereby forming immunoproteasomes. The immunoproteasomes are capable of cleaving other substrates and consequently have other functions than standard proteasomes. For a long time they have only been implicated in antigen presentation. Immunoproteasomes were found to be more efficient in the generation of antigenic peptides that can bind MHC class I molecules. After transport to the cell membrane these peptide-MHC complexes induce effective immune responses. However, we know now that processing and presentation of several antigenic peptides requires the presence of the standard proteasome. In presence of immunoproteasomes internal destructive cleavage of these peptides occurs, which prevents the presentation of the peptide. Recently, immunoproteasomes have also been implicated in other immune-related processes, including the production of cytokines. Interesting though is the expression of immunoproteasomes in non-immune cells and in immune-privileged tissues like the eyes and brain, suggesting a function beyond immunity. Indeed, this type of proteasomes was found to be more efficient in the degradation of poly-ubiquitylated proteins and accumulating proteins upon stress signals and thereby in the maintenance of cell viability. Which of the immune or non-immune functions is executed by the immunoproteasomes might be dependent on the cell type expressing these proteasomes. However, more knowledge is required to understand the exact mechanisms of immunoproteasome functioning. This information might furthermore help in the development of new treatments for a variety of diseases, as many diseases are associated with (immuno)proteasome disfunctioning. Lione Willems | Immunoproteasomes and their functions in immunity and cell viability 20