* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PPT - NIH LINCS Program
Survey
Document related concepts
Transcript
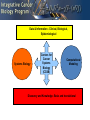
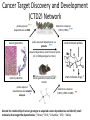
NCI Projects Relevant to LINCS Jennifer Couch, Ph.D. Daniela Gerhard, Ph.D. National Cancer Institute, NIH A Long History of Using Cell Lines to study disease: NCI 60 • • • • • • • Genes and Genetics Complex Signaling Networks Multiple Cellular Processes Microenvironment Host Systems Environmental Factors Population Dynamics Initiation Progression Time - Progression Metastasis Recurrence Histology Variation Cancer is Complicated The ICBP Approach Data & Information - Clinical, Biological, Epidemiological Systems Biology Centers for Cancer Systems Biology (CCSB) Computational Modeling Discovery and Knowledge- Basic and translational ICBP2:Centers for Cancer Systems Biology Cancer Stage Sander Califano Plevritis Cell – cancer differentiation; single cell analysis; disease progression and resistance Leukemia, Lymphoma Multiscale analysis of Genomic and Cell Networks (MAGNet), B cell lymphoma, glioma Golub Integrate multiple data types to identify essential genenew drug targets Lung Melanoma Lauffenburger Quantative modeling of critical cancer process; growth, migration, DNA repair Brain, lymphoma, Breast Quaranta 3-D model cell-cell communication; microenvironment; variability of drug response Melanoma, breast, panceas, glio, CLL Cellular model of tumor heterogeneity; cell phenotype measurements; multiscale Breast, Lung Clarke Hormone responsiveness; population exposure risk; GWAS; drug design Breast Friend Cross disease analysis; Sage Bionetworks; network data integration Glioma, Ovarian, colon, Liver, Meduloblastoma, Pancreas, Breast, ect Wong Tumor Stem cell, imaging, ME interations; protein cDNA array Breast Hlatky Tumor evolution; quanatative analysis; cellular population dynamic modeling Prostate, Breast Gray Modeling Approach Cancer heterogeneity; molecular signaling and combinational therapy Breast Huang Epigenetic and microRNA examination of hormonal –chemo resistance Prostate, Ovarian Breast Physical Scale Lawrence Berkeley Labs (Gray) Model Based Predictions of Responses to RTK Targeted Therapies in Breast Cancer Overall: Infer signaling network for cancer subtypes; model response to MAPK inhibitors, Her2 targeted therapies and P13K targeted therapies Project 3 :HER-family signaling deregulated in breast cancer Glucose • Therapeutic perturbations of a breast cancer cell line panel show: GF GLUT4 RTK PDK1 Glucose AS160 PI3K SHP2/ SHC/SOS/ GRB2 C-Src HK2 Ras G6P Stat3/5 Rac AKT ATP AMP F6P Rho Raf LKB1 PFK1 cdc42 MEK1,2 SGK3 GADP F1-6BP AMPK MAPK1,2 GSK3b 1,3 BPG MEKK4 MEKK6 mTOR 3PG MEK4 MEK6 JNK/ SAPK p38 PK-M2 2PG PEP HIF1a Pyruvate LDH PDH RSK TSC2 Rheb PDHK1 BAD AcetylCoA Lactate FOXO3A p70S6K GS TCA-Cycle S6 ER ACL Glycogen synthesis Citrate AcetylCoA MalonylC oA FAS CREB Fos ACC RB Jun ER ATF2 AP1 Glycolysis Fatty Acid Synthesis Gluconeog enesis Cellular respiration Stat3/5 c-Myc CBX5 • Pathway function and mechanism of deregulation differ according to subtype • Responses are subtype specific • Responses are not durable and mechanisms are not understood • Therapeutic responses depend on the chemical and mechanical microenvironment ICBP: Efforts to Expand the Field • Community resources – Software tools and models – Data sharing/Data portal – Biological (ICBP 43 cell lines) • Educational /Outreach – Undergraduate opportunities – Junior PI training and mentoring – Curricula development • Meetings/ workshop – – – – – PI meetings Joint meetings (other consortia, etc.) Junior Investigator meeting (yearly) Mathematical Cancer Modeling workshop (2010, 2012) National Cancer Systems Biology meeting/ AACR (March 2011) ICBP 43 http://physics.cancer.gov/about/ Physical Sciences-Oncology Centers (PS-OCs) Network: Cel Line Project • Selected human immortalized cell lines – MCF-10A: non-malignant breast epithelial cells – MDA-MB-231: metastatic breast cancer cells • Scope of project was to have each PS-OC conduct their unique physical science measurements on the cell lines and share and cross-compare datasets • Cell lines were propagated by one PS-OC Investigator and distributed to one site at each of the 12 PS-OCs along with detailed SOP for cell culture of each cell line • Requirement to upload cell line data to a pilot data coordination site • Investigators presented results at Data Jamboree Meeting in January 2011 • Cell line project data will serve as a pilot dataset for building the PS-OC Data Coordination Center (DCC) • Currently a Network manuscript is in preparation describing the results of the project Cell Line Project Physical Science Measurements: Molecules to Cells Genomics Cornell • Gene expression analysis in response to changes in pO2, pH and metabolic load in 3D alginate MIT • Single-cell transcript counting NU • TEM and cryo-EM analysis of chromatin structure and distribution of chromatin-organizing proteins • Light scattering and TEM roughness nanoscale measurements • Deep sequencing-based nucleosome position analysis •Metaphase chromosome mechanics • Magnetic tweezer analysis of chromatin polymer elasticity •DNA replication dynamics analysis Princeton • Analysis of chromatin and mitochondria as a function of time and position on stressed landscapes •Genomic analysis of cells transported to and from stressed landscapes Scripps • Nuclear dimensions (radius, aspect ratio) Proteomics NU • Proteasome processing analysis USC • SILAC for MSbased proteomics Cell Mechanics/ Morphology ASU • Cell CT • AFM JHU • Cadherins via single- molecule force microscopy (2D/3D) • Ballistic Intracellular Nanorheology (BIN; 2D/3D) • Intracellular microrheology for cells in 2D & 3D MIT • Cell mass measurements during cell cycle Cell Surface/ Adhesion Cornell • FACS for adhesion receptors • Adhesion assay to E-selectin surfaces under shear stress • Aggregation with human leukocytes under flow JHU • Subcellular release assay to measure rate of de-adhesion • Flow chamber assay to measure cellmatrix adhesion strength Moffitt • Colony size/morphology in response to altered pH and pO2 • Cell trace Princeton • Cell morphology as a function of time and position on stressed landscapes Scripps • Cell dimensions (radius, aspect ratio) • Cortical tension • Cytoplasmic viscosity •Elasticity (E) • Cellular complexity/granularity UCB • ECM stiffness effects on cell polarity and spreading Princeton • Analysis of cell adhesion as a function of time and position on stressed landscapes under flow conditions Methodist • In vitro nanoparticle •internatlization The Cancer Target Discovery and Development (CTD2) Network LINCS Consortium Kick-Off Meeting October 28, 2011 Daniela S. Gerhard, Ph.D. Director, Office of Cancer Genomics Large Projects Examples of NIH Investment in Genomic Research • Therapeutically Applicable Research to Generate Effective Treatment (TARGET) • The Cancer Genome Atlas (TCGA) • Cancer Genome Anatomy Project/Cancer Genome Characterization Initiative (CGAP/CGCI) • Genome-wide association studies (GWAS) of common and complex diseases and follow-up (~60/450 grants are cancerrelated) Data generated is made publicly available • ~20% of NIH ARRA funded genomic projects Molecular Characterization of Cancer Tissues is Essential but not Sufficient • Each tumor has hundreds to thousands genomic alterations – Chromosomal changes: amplifications, deletions, translocations – Epigenetic changes – Mutations • Little is known about the cellular function of most genes, much less how sequence variants and mutations affect them – Distinguishing initiating vs. driver vs. passenger mutations • Drivers are defined as genes involved in tumor maintenance • Evidence is accumulating that multiple subclones exist within a tumor and their frequency varies between patients • As tumors evolve genes essential for survival may be different from those that were necessary early on – Genomic alterations result in cancer within specific context • Cell of origin • Other molecular alterations in genes that may have synergistic or antagonistic impact ARRA Opportunity Question: Can a network be formed that would effectively address a current major scientific challenge: efficient transition from patient-based large multi-dimensional genomic data target validation small molecule modulators (therapy, not part of the initiative) How to advantage the flood of genomic data and accelerate the transition to treatments of patients based on the genomic profile of their cancer? ARRA Cancer Target Discovery and Development (CTD2) Network Centers • Broad Institute, Cambridge, Massachusetts PI: Stuart Schreiber, Ph.D. • Cold Spring Harbor Laboratory, Long Island, New York PI: Scott Powers, Ph.D., co-PI: Scott Lowe, Ph.D. • Columbia University, New York, New York PI: Andrea Califano, Ph.D. • Dana-Farber Cancer Institute, Boston, Massachusetts PIs: William Hahn, M.D., Ph.D., L. Chin, M.D. and R. DePinho, M.D. • University of Texas Southwestern Medical Center, Dallas, Texas PI: Michael Roth, Ph.D., co-PIs: M. White, Ph.D., J. Minna, M.D. http://ocg.cancer.gov/programs/ctdd.asp ARRA CTD2 Network • Each application included up to 3 mature projects • Functional network formed rapidly – Component centers share results “in real time” (precompetitive) • Established an ethos of data and resource sharing with scientific community upon validation – IT WG developed file formats for data sharing compatible with Cancer Data Standards Registry and Repository (caDSR) within caBIG • Enabled experiments, using new data generated by the molecular characterization projects to identify candidate targets, small molecule modulators and mechanisms: one example was ovarian cancer Cancer Target Discovery and Development (CTD2) Network probe acquired *** dependencies via RNA determine relevance (STK33; TBK1) *** probe acquired dependencies via proteins *** cancer genomics small-molecule probes (acquired dependency small-molecule probe kit in >400 genotyped cell lines) cancer patients probe acquired dependencies via network analyses * cancer genomics-based mouse ** models small-molecule drugs determine relevance (STAT3; C/EBP in GBM) ** Decode the relationship of cancer genotype to acquired cancer dependencies and identify small molecules that target the dependencies (*Broad; *CSHL; *Columbia; *DFCI; *Dallas) * Summary of the CTD2 Network’s caOv Results The power of the network: made rapid progress by sharing data, working together and taking advantage of complementary, non-overlapping expertise to carry out the experiments. Each Center contributed: – Identified candidate signature to stratify patients into best and worst prognostic groups – Identified candidate targets for therapeutic development • Confirmed a subset of candidates by in vitro and ex vivo experiments – Identified candidate small molecules for a subset of confirmed targets – Plan to generate mouse models for in vivo screening of other candidate genes within a specific genetic context – Experiments are ongoing Critical lesson: collaborative efforts to integrate several methods can yield exponential gains relative to the incremental gains achieved through improving any single method (united they are more than a sum of parts) CTD2 Network Research Mission • Shift current research paradigms in translation pathway of patient-derived multidimensional genetic data to the clinic and utilize novel concepts, approaches and methodologies • Develop research that will exert a sustained influence on the field • Develop a pre-competitive culture to ensure sharing of data, methods (analytical, experimental) and reagents within the network and the scientific community at large Goals for the New Network • Accelerate the translation of patient genomic data into clinical application – Innovate the integration of computational mining large scale genomic data analyses • Make tools available through web – Identify and confirm new therapeutic target candidates – Identify and confirm novel modulators within specific cancer context (cellular or mutational) in vitro (cell lines) or in vivo (cancer models) • Small, stereochemically “interesting“ molecules – Use of novel organist chemistry – molecules more “natural products-like” – Mature molecules: optimize activity, structure activity relationship, systematic variation of stereochemistry • siRNAs – Multi-expertise team – Share models and reagents with the scientific community – Share data and methods with the scientific community through the web • As genomic data become available from TARGET, TCGA etc.,: be nimble, flexible and open to new opportunities The Cancer Target Discovery and Development (CTD2) Network LINCS Consortium Kick-Off Meeting October 28, 2011 Daniela S. Gerhard, Ph.D. Director, Office of Cancer Genomics