* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Transpeptidase, or PBP
Cre-Lox recombination wikipedia , lookup
Nicotinic acid adenine dinucleotide phosphate wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of genetic engineering wikipedia , lookup
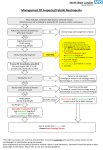
Mechanisms of antimicrobial action directed against the bacterial cell wall and corresponding resistance mechanisms M-4 Advanced Therapeutics Course Mechanisms of antimicrobial resistance Drug-modifying enzymes (e.g., - lactamases, aminoglycosidemodifying enzymes) Altered drug targets (e.g., PBPs ribosomes, DNA gyrase) Altered uptake or accumulation of drug (e.g., altered porins, membrane efflux pumps) Subunits for cell wall construction N-acetylglucosamine N-acetylmuramic acid pentapeptide D-ala-D-ala Cell Wall Assembly Second layer of cell wall cross-linked to the lower layer Layer of cell wall with cross links of 5 glycines (gray) A subunit is added to the growing chain Transpeptidase (PBP) forms a 5-glycine bridge between peptides Transpeptidase, or PBP (orange sunburst) is bound by beta-lactam antibiotic (light blue) and its activity is inhibited (turns gray) 5-glycine crosslinking bridges cannot form in the presence of a beta-lactam, and the cell wall is deformed and weakened Mechanisms of beta-lactam resistance • Drug-modifying enzymes (beta-lactamases) – Gram-positives(e.g., S. aureus) excrete the enzyme – Gram-negative (e.g., E. coli) retain the enzyme in the periplasm • Overexpression of • cell wall synthetic enzymes – e.g., vancomycin-intermediate S. aureus (VISA) Alteration of the PBPs so antibiotic cannot bind – e.g., S. pneumoniae, gonococcus • Exclusion from the site of cell wall synthesis – Porin mutations in the outer membrane of Gramnegative bacteria only (e.g., Ps. aeruginosa) Beta-lactamases Beta-lactamases (dark orange) bind to the antibiotics (light blue) and cleave the beta-lactam ring. The antibiotic is no longer able to inhibit the function of PBP (orange sunburst) Beta-lactamase activity Altered drug targets Vancomycin-intermediate S. aureus Production of excessive cell wall; the antibiotic cannot keep up MRSA vancomycin MIC = 2 µg/ml VISA vancomycin MIC =8 µg/ml MRSA VISA Mechanism of vancomycin action V D-ala-D-ala Mechanism of vancomycin resistance Vancomycin is unable to bind to the D-ala-Dlactate structure D-ala-D-lactate V ·June 2002: isolated from the catheter exit site in a chronic dialysis patient ·The patient had received multiple courses of abx since April 2001; toe amputation in April 2002 --> MRSA bacteremia ·VRSA also found at amputation stump wound (with VRE and Klebsiella); not in the patient’s nose ·Vancomycin MIC >128mcg/ml!! (contains vanA) ·Sensitive to trim/sulfa, chloro, tetracyclines, Synercid, linezolid MRSA and penicillin-resistant S. pneumoniae • These bacteria are both resistant because they • • have altered bacterial targets -- penicillin-binding proteins (PBPs or transpeptidases) In MRSA, the altered PBP2 (mecA) gene is acquired by gene transfer from another bacterium. In pneumococci, the alteration in PBP is generated by uptake of DNA released by dead oral streptococci and recombination at the pneumococcal pbp gene to create a new, chimeric protein that does not bind penicillin. – depicted on the next slide . . . DNA alpha-strep pbp Alpha-strep S. pneumoniae transformation alpha-strep pbp S. pneumoniae chromosomal pbp; penicillin-sensitive Chimeric pbp (resistant to penicillin) Outer membrane permeability in Gram-negative bacteria Beta-lactam (blue) enters through an outer membrane porin channel Altered porin channel prevents access of the antibiotic to the cell wall Outer membrane Cell wall (peptidoglycan) Inner membrane Cytoplasm Bacterium