* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download allergen
Cell-penetrating peptide wikipedia , lookup
Epitranscriptome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein–protein interaction wikipedia , lookup
DNA vaccination wikipedia , lookup
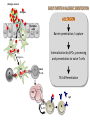
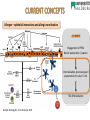
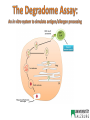
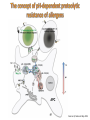
Fátima Ferreira Department of Molecular Biology University of Salzburg, Austria • Derive from a variety of environmental sources • Display a broad range of structures and functions in their respective sources (e.g. structural, enzymatic, ligand-binding) • Represent a small fraction of proteins that humans are exposed to • The same proteins behave as allergens across the human population • Natural exposure: allergens are delivered as complex mixtures of molecules How do such diverse proteins preferentially induce Th2-biased immune responses? Allergen source Allergen Epithelial cells Barrier penetration / capture DC DC Internalization by APCs, processing and presentation to naïve T cells Migration DC T B Naïve T cell Th2 differentiation Th2 IgE IL-4 IL-13 B B Allergen – epithelial interactions and allergic sensitization Engagement of PRRs Barrier penetration / capture Internalization, processing and presentation to naïve T cells Th2 differentiation Gandhi & Vliagoftis, Front Immunol 2015 Post-translational modifications • Glycosylation • Disulfide bonds • Proteolytic processing • Chemical modifications (nitration, deamidation) Biochemical Function • Enzyme activity (protease) • Ligand-binding (lipids, ions) • Receptor binding/activation (PARs, TLR, MR, CLR) Oligomerization • Dimers • Oligomers Protein Fold and Stability • Molecular dynamics • pH • Temperature • Proteolytic susceptibility of the antigen 1. Preparation of endolysosomal fraction 2. In vitro degradation (pH 4.5) - Intact protein + DCs APC-derived endolysosomal proteases 3. Analysis SDS-PAGE/ Scanner densitometry Antigen/Allergen Homogenization Centrifugation 0h 0.5h 1h Post-nuclear supernatant - Peptide sequencing (MS/MS) 2h Ultracentrifugation 3h Capillary HPLC/ESIQTOF Mass Spectrometry 4h Microsomal fraction Freeze-thaw cycles Endolysosomal Proteins/proteases .. .. .. .. 5h 48h Egger et al, PLoS One 2011 Bet v 1.0401 (Bet v 1d) Bet v 1.0101 (Bet v 1a) activates DCs much better (elevated differs from Bet v 1.0401 by 7 levels of CD80 and CD86; reduced amino acids (sequence homology of IL-6 production) and induces 95%) stronger IgG and IgA responses in mice3 Bet v 1d Bet v 1a Bet v 1d Zaborsky et al (2010). J Immunol 184: 725-735 Bet v 1a Egger et al, PLoS One 2011 Natural Pru p 3 (nPru p 3) Reduced and alkylated Pru p 3 (R/A Pru p 3) Toda et al., JACI 2011 Natural Pru p 3 (nPru p 3) Degradome ½ life: Approx. 12 hours Reduced and alkylated Pru p 3 (R/A Pru p 3) Degradome ½ life: approx. 2 hours Toda et al., JACI 2011 LIGAND-BINDING NITRATION Ackaert et al., PlosOne 2014 Kofler et al., J Mol Biol 2012; Asam et al., CEA 2014 Grutsch et al., Biophys J 2014 DIMERIZATION FOLD STABILITY Zaborsky et al., J Immunol 2010; Egger et al., PlosOne 2011 Kofler et al., J Biol Chem 2014 Wallner et al., JACI 2011; Machado et al., JACI 2015 Wallner et al. (in preparation) Bet v 1d (dimer) Bet v 1a (monomer) Bet v 1a T. Thermophilus homologue POST-TRANSLATIONAL MODIFICATIONS (disulfide bridges) Toda et al., JACI 2011 Susceptibility/resistance to endolysosomal proteases seem to be linked to “intrinsic” allergenicity/immunogenicity of protein antigens and influences the type of immune response (Th2 vs. Th1 bias) Proteolytic resistance Delamarre, L et al (2005) Science 307: 1630-1634 Delamarre, L et al (2006) J Exp Med 203: 2049-2055 Egger et al, PLoS One 2011 pH 4.5 pH 5.0 Bet v 1a Bet v 1d Egger et al, PLoS One 2011 Freier et al, Nature Sci Rep 2015 SAW-measurements pH 6.7 Freier et al, Nature Sci Rep 2015 Freier et al, Nature Sci Rep 2015 The interplay between fold stability and pHdependent proteolytic susceptibility… Fold stability IgE reactivity pH and proteolytic susceptibility Immunogenicity/allergenicity depends on differential fold stability at different endosomal pH In vivo immunogenicity/allergenicity IgG1 IgG2a IL-4 IFN-g RBL IgE Machado et al., JACI 2015 Machado et al., JACI 2015 Chemical modifications Post-translational modifications ALLERGEN ENGINEERING (HYPOALLERGENS) RECOMBINANT ALLERGY VACCINES Ligand-binding Oligomerization Machado et al., JACI 2015 • Susceptibility/resistance to lysosomal proteases seem to be linked to intrinsic allergenicity/immunogenicity of protein antigens and to influence the type of immune response (allergic vs. non-allergic) • Differential pH-dependent fold stability along endosomal maturation (acidification) seem to be an important factor for allergenicity and immunogenicity • Fine tuning of resistance/susceptibility to endolysosomal proteolysis may be an approach for designing allergen vaccines with enhanced immunogenicity Research Assistants Michael Wallner PhD students Gabi Gadermaier Lorenz Aglas Stephanie Eichhorn Isabel Pablos Sabrina Schuller Anargyros Roulias Heidi Hofer Teresa Stemeseder Claudia Asam Sara Huber Martin Wolf Markus Steiner Olivia McKenna Josef Thalhamer Peter Lackner Hans Brandstetter Peter Briza Albert Duschl Christian Huber Yoan Machado Gernot Achatz Barbara Bohle Beatrice Jahn-Schmidt Heimo Breiteneder Karin Hoffmann-Sommergruber Christof Ebner Martin Tollinger Klaus Liedl Adriano Mari Ronald van Ree Program BioScience & Health Ana Paula Valente Fabio Almeida (Uni Salzburg)