* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download my_biological_databses
Non-coding DNA wikipedia , lookup
Gene regulatory network wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene expression wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Community fingerprinting wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Homology modeling wikipedia , lookup
Genome evolution wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
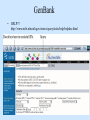
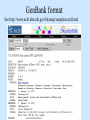
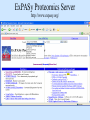
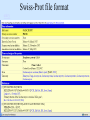
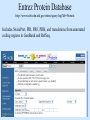
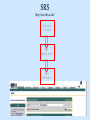
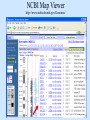
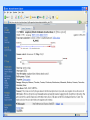
Biological databases International nucleotide sequence Database collaboration. DDBJ (Japan) EMBL European Molecular Biology Laboratory http://www.ebi.ac.uk PubMed, GenBank Nucleotides (NCBI) Proteins http://www.ncbi.nlm.nih.gov Genomes Taxonomy Structure Domains NCBI - GenBank • GenBank: All publicly available nucleotide and amino acid sequences. • Data Source: 1. 2. 3. • Direct submission from scientists Literature. Genome Sequencing DNA database divisions (examples) 1. 2. 3. Organism division (Human, Bacteria, etc). Molecule division (DNA, RNA, protein). Sequence division (Genome, ESTs STSs). sequence databases An optimal database should be: Comprehensive, well annotated, easily searched & easy data retrieval, provide cross-references The GenBank database: As of April 2004, there are over 8,989,342,565 bases in GenBank. Problems 1: huge databases Redundancy and inadequate sequences. Problem 2: Submission by users Redundancy, Only the submitter can change it, not always up to date, partial annotation. GenBank • HELP!!! http://www.ncbi.nlm.nih.gov/entrez/query/static/help/helpdoc.html Unique Identifiers at NCBI accession numbers apply to a complete sequence record sequence identification numbers apply to the individual sequences within a record GI number assigned consecutively by NCBI to each sequence it processes Version number accession number followed by a dot and a version number. •The format of accession numbers varies, depending upon the source database: •GenBank/EMBL/DDBJ - One letter followed by five digits, e.g.: U12345 or two letters followed by six digits, e.g.:AY123456 •Swiss-Prot - All are six characters: [O,P,Q][0-9][A-Z,0-9][A-Z,0-9][A-Z,0-9][0-9] e.g.:P12345 and Q9JJS7 •RefSeq - Two letters, an underscore bar, and six digits, e.g.:NM_000492 (mRNA) NT_ (contig) NC (chromosome) NG (genomic region). • If a sequence changes in any way, it receives a new GI number, and the version number is incremented by one. GenBank format See http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html GenBank format FASTA format Example: >my_sequence_name BTYKLJGJFKHVHFMGHF KHGJFJFVKHGJHLNLNLJ KJGKGKGKHLJH • Easy to parse • Least informative • Default input format for sequence analysis software (e.g., BLAST, CLASTALW). Swiss-Prot (http://www.ebi.ac.uk/swissprot/) • Core data: sequence, taxonomy and bibliographic reference. • Annotation data: function, domain structure, post-translational modifications, protein variants, etc. – a curated protein sequence database – provide a high level of annotation – minimal level of redundancy – high level of integration with other databases (cross references). TrEMBL • a computer-annotated supplement of Swiss-Prot that contains all the translations of EMBL nucleotide sequence entries not yet integrated in Swiss-Prot. ExPASy Proteomics Server http://www.expasy.org/ Swiss-Prot file format entry Flat-file original Swiss-Prot format Search sequence databases Two search methods – Text based searching– searches textual information contained in header sections of database entries – Sequence search– searches sequence information with sequence queries – next week! Text based searching - Search for query words in specific fields. - Choose your database and add limits. - Examples: Entrez, SRS. NCBI – Entrez (http://www.ncbi.nih.gov/Entrez/) • Entrez is the search tool for NCBI databases. • The search starts by choosing the relevant group of databases (Nucleotide, Protein, etc). • Use field qualifiers, logical operators, and a “limits” form. • Boolean operator, AND, OR, NOT Group together by using () Example: cytochrome AND human cytochrome AND (human OR mouse) • Always use upper case for operators. • If you don’t use any operator the query words are looked together! • Field qualifiers: Search in the specific field: Author, organism, journal … Example: • homo sapiens [organism] AND kinase AND nature [journal] • Cytochrome b • Cytochrome b AND human • Cytochrome b AND human[organism] • Cytochrome b AND human[organism] and limits. Entrez Protein Database http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein Includes SwissProt, PIR, PRF, PDB, and translations from annotated coding regions in GenBank and RefSeq. Entrez Nucleotides database http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide • Includes GenBank, RefSeq, and PDB. • As of April 2004, there are over 38,989,342,565 bases. SRS http://srs.ebi.ac.uk/ Choose Library Fill Query form Get Results Gene-centric Databases • Repository-type database: - Many pieces of sequences related to a sequence - Examples: GenBank/SwissProt • Gene-centric database: - All the sequence information relevant to a given gene is made accessible at once: Get the whole story at once! - Provide easy access when the query is related to a gene or function. - Examples: Gene, UniGene, RefSeq. Gene http://www.ncbi.nih.gov/entrez/query.fcgi?db=gene • Gene provides a unified query environment for genes • Query on names, symbols, accessions, publications, GO terms, chromosome numbers, E.C. numbers, and many other attributes associated with genes and the products they encode. • Unique identifiers assigned to genes with known map positions. • Supply key connections of map, sequence, expression, structure, function, citation, and homology data. • Provide identifiers to UniGene, RefSeq, relevant GenBank entries, OMIM and SNPs. • Can be considered as the successor to LocusLink Refseq http://www.ncbi.nlm.nih.gov/projects/RefSeq/ • non-redundancy • distinct accession series • updates to reflect current knowledge of sequence data and biology • ongoing curation by NCBI staff and collaborators, with reviewed records indicated. • data validation and format consistency ESTs division Uses: 1. 2. 3. Gene predication. Expression level (only clues). Alternative splicing. Problems: 1. 2. 3. Redundant database. mistakes (single read-through). Incomplete coverage of genes: - Only for Model eukaryotic organisms Rare tissues Low copy number of genes UniGene http://www.ncbi.nlm.nih.gov/UniGene • An automatically partitioning of GenBank sequences into a non-redundant set of gene-oriented clusters. • Each UniGene cluster contains sequences that represent a unique gene, as well as related information such as the tissue types in which the gene has been expressed and map location. • Focus on mRNA and EST information Wouldn’t it be great if… Annotation Tracks sequence Genome backbone: base position number chromosome band sts sites gap locations known genes predicted genes microarray/expression data evolutionary conservation SNPs repeated regions more… Links out to more data Solution: Genome Browsers, Or “map Viewers” NCBI Map Viewer http://www.ncbi.nlm.nih.gov/Genomes/ Ensemble (http://www.ensembl.org/) • Ensemble example: http://www.ensembl.org/Docs/linked_docs/human_eg_19_34.pdf UCSC Home page ( genome.ucsc.edu ) navigate General information navigate Specific information— new features, current status, etc. UCSC Material developed by W.C. Lathe and M. Mangan, [email protected]