* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Amino acid
Nucleic acid analogue wikipedia , lookup
Western blot wikipedia , lookup
Catalytic triad wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Peptide synthesis wikipedia , lookup
Structural alignment wikipedia , lookup
Point mutation wikipedia , lookup
Homology modeling wikipedia , lookup
Genetic code wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Biosynthesis wikipedia , lookup
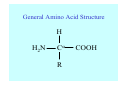
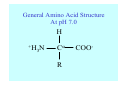
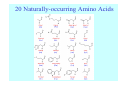
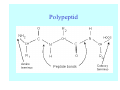
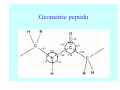
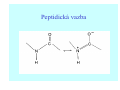
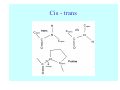
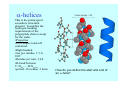
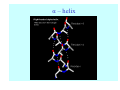
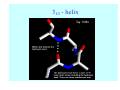
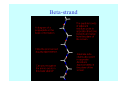
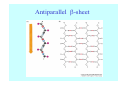
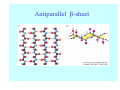
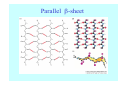
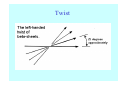
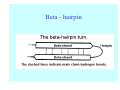
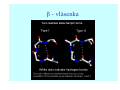
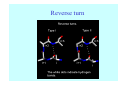
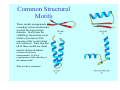
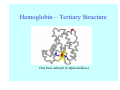
Struktura proteinů • • • • • Aminokyseliny Primární Sekundární Terciární (Geometrie) Kvarterní • Domény, Motivy Struktura proteinů • • • • • Hlavní řetězec Vedlejší (boční) řetězce Het – grupy (hem, Fe4S4 ,Zn2+) Sacharidy Solvent Protein Function in Cell 1. Enzymes • Catalyze biological reactions 2. Structural role • • • Cell wall Cell membrane Cytoplasm General Amino Acid Structure H H2N α C R COOH General Amino Acid Structure At pH 7.0 H +H N 3 α C R COO- General Amino Acid Structure Atom numbering Cα, Cβ, Cγ, Cδ, Nε, Cζ, Nκ CA, CB, CG,CD, NE,CZ, NK A: -CB I: -CB(CG1)-CG2-CD R: -CB-CG-CD-NE-CZ(NK1)-NK2 Chirality: Glyceraldehyde D-glyderaldehyde L-glyderaldehyde CORN 20 Naturally-occurring Amino Acids Single and 3-Letter Codes • • • • • • • • • • Alanine Aspartic AciD Phenylalanine Histidine Lysine Methionine Proline ARginine Threonine Tryptophan Ala A Asp D Phe F His H Lys K Met M Pro P Arg R Thr T Trp W Cysteine Glutamic Acid Glycine Isoleucine Leucine AsparagiNe Glutamine Serine Valine TYrosine Cys C Glu E Gly G Ile I Leu L Asn N Gln Q Ser S Val V Tyr Y Properties of the amino acids aliphatic and aromatic Side chain pKa Amino acid Occurrence Molecular weighta Volume Å3 Aliphatic Glycine Gly G 7.2 57.05 48 Alanine Ala A 7.8 71.09 67 Valine Val V 6.6 99.14 105 Leucine Leu L 9.1 113.6 124 Isoleucine Ile I 5.3 113.6 124 Proline Pro P 5.2 97.12 90 Phenylalanine Phe F 3.9 147.18 135 Tyrosine Tyr Y 3.2 163.18 141 Tryptophan Trp W 1.4 186.21 163 Aromatic aMolecular 10.5 weight of nonionized amino acid minus that of water. Properties of the amino acids polar-uncharged and charged Side chain pKa Amino acid Occurrence Molecular weighta Vol. Å3 Polar uncharged Serine Ser S ~13 6.8 87.08 73 Threonine Thr T ~13 5.9 101.11 93 Cysteine Cys C 8.4 1.9 103.15 86 Methionine Met M 2.2 131.19 124 Asparagine Asn N 4.3 114.11 96 Glutamine Gln Q 4.3 128.14 114 Lysine Lys K 10.5 5.9 128.17 135 Arginine Arg R 12.5 5.1 156.19 148 Histidine His H 6.0 2.3 137.14 118 Aspartate Asp D 3.9 5.3 115.09 91 Glutamate Glu E 4.1 6.3 129.12 109 Positively Charged Negatively Charged aMolecular weight of nonionized amino acid minus that of water. Polypeptid Peptide Bond • • The peptide bond influences all aspects of protein structure and function. Key features: – 1. Planar – 2. Fairly rigid, due to partial double bond character. – 3. Almost always in trans configuration. – 4. Polar. Can form at least two hydrogen bonds. – 5. Places restrictions on the conformation of the polypeptide chain. Geometrie peptidu Peptidická vazba Cis - trans Chemické modifikace Syntéza Genetický kód Acetylace terminální aminoskupiny Odštěpování částí peptidů (insulin) Hydroxylace prolinu (hydroxyprolin, Hyp) Karboxylace glutamové kyseliny Fosforylace (serin, tyrozin) Glykoproteiny (glukóza, galaktóza) Zelené proteiny (medúza) Disulfide Bonds • Side chain of cysteine contains highly reactive thiol group • Two thiol groups form a disulfide bond Disulfide Bridge Disulfide Bridge – Linking Distant Amino Acids Protein Structure Protein Structure Hemoglobin – Primary Structure NH2-Val-His-Leu-Thr-Pro-Glu-Glu- Lys-Ser-Ala-Val-Thr-Ala-Leu-TrpGly-Lys-Val-Asn-Val-Asp-Glu-ValGly-Gly-Glu-….. beta subunit amino acid sequence Konformace Bond Rotation Determines Protein Folding G.N. Ramachandran • Used computer models of small polypeptides to systematically vary φ and ψ with the objective of finding stable conformations • For each conformation, the structure was examined for close contacts between atoms • Atoms were treated as hard spheres with dimensions corresponding to their van der Waals radii • Therefore, φ and ψ angles which cause spheres to collide correspond to sterically disallowed conformations of the polypeptide backbone Computed Ramachandran Plot White = sterically disallowed conformations (atoms come closer than sum of van der Waals radii) Blue = sterically allowed conformations Ramachandran Plot Experimental Ramachandran Plot φ, ψ distribution in 42 high-resolution protein structures (x-ray crystallography) Specificita Aa Alanine Ramachandran Plot Glycine Ramachandran Plot Note more allowed regions due to less steric hindrance - Turns Proline Ramachandran Plot Note less allowed regions due to structure rigidity Secondary Structural Elements • • • • α-helices β-sheets Turns Random coil φ, ψ and Secondary Structure Name φ ψ Structure ------------------- ------- ------- --------------------------------αR -57 -47 right-handed alpha helix. αL +57 +47 left-handed alpha helix . 310 -49 -26 right-handed helix. π -57 -80 right-handed helix. Type II -79 150 left-handed helices formed by polyglycine and polyproline. Collagen -51 153 right-handed coil formed of three left handed helicies. Ramachandran Plot and Secondary Structure • Repeating values of φ and ψ along the chain result in regular structure • For example, repeating values of φ ~ -57° and ψ ~ -47° give a right-handed helical fold (the alphahelix) The structure of cytochrome C shows many segments of helix and the Ramachandran plot shows a tight grouping of φ, ψ angles near -50,-50 alpha-helix cytochrome C Ramachandran plot Similarly, repetitive values in the region of φ = -110 to –140 and ψ = +110 to +135 give beta sheets. The structure of plastocyanin is composed mostly of beta sheets; the Ramachandran plot shows values in the –110, +130 region: beta-sheet plastocyanin Ramachandran plot Konformace – „all trans“ α-helices This is the prototypical secondary structural element. It satisfies the hydrogen bonding requirements of the polypeptide chain (except for the ends). •Properties •It is compact and self contained. •Right handed •rise per residue, 1.5 Å, 100o •Residue per turn, 3.6Å •H-bond between C=O(n).... .H-N(n+4) •period -18 residue, 5 turns How do you define the start and end of an α-helix? α – helix R-groups are on outside of α-helix Certain amino acids are "preferred" & others are rare in α−helices • Ala, Glu, Leu, Met = good helix formers • Pro, Gly Tyr, Ser = very poor Obecné spirály n/m n - počet residue na otočku m - počet atomů v cyklu H-vazby 2.27 310 G 3.613 H αR 4.416 π 310 - helix Beta-strand β-sheets β-sheets fulfill the hydrogen bonding potential of the main-chain atoms, except at the edges. Adjacent strands are usually close in sequence. Properties: Distance between CA's is ~3.6 Å in an extended strand Distance between strands ~4.6 Å Strands are not flat. They have a characteristic righthanded twist Antiparallel β-sheet Antiparallel β-sheet Parallel β-sheet Mixed β-Sheets also occur Topologie Twist Turns Turns are a major structural component of proteins. Without them there could be no globular proteins. They are characterized by an irregular series of conformational angles that fold the chain back on itself. Turns are often very compact and well ordered, though they are hot-spots for evolution. Sometimes they are sites of flexibility, at other times they are quite rigid. Need to look carefully at the structure. Turns can be classified into a few well defined arrangements. Beta - hairpin β - vlásenka Beta-hairpin Reverse turn Reverse turn - Rama Tertiary Structure • All alpha • All beta • Alpha+beta (mix) • Alpha/beta (separated) Common Structural Motifs These simple arrangements of secondary structural elements account for most protein domains. In all cases the stabilizing interactions occur within a local area of the sequence (this is convenient for evolution). Note also that all of these motifs are chiral and are observed almost exclusively in these arrangements. Is this a consequence of the chirality of the amino acids? Why are these common? Domains • A compact, spatially distinct unit • Other definitions – A region of a protein with an experimentally assigned function – A region of a structure that recurs in different contexts in different proteins – A region with a separate hydrophobic core – … – Or a combination of these Quaternary Structure • Multiple chains form a protein – collaborate to perform a function Major secondary structural elements of HIV-1 protease The secondary structure is mostly βstrand and random coil. There is one small helix and two β-turns. The β-sheets show their characteristic twist and associate into two layers, which is common in βstructures. The structure is compact, though this is not apparent in these ribbon representations. Space Filling Representation of HIV-1 Protease Ribbon representations are convenient for showing the path of the polypeptide chain but are misleading with respect to the compactness. What is not evident in this representation? Hydrogens? Water? Dynamic properties? Hemoglobin: Background • Protein in red blood cells • Composed of four subunits, each containing a heme group: a ring-like structure with a central iron atom that binds oxygen • Picks up oxygen in lungs, releases it in peripheral tissues (e.g. muscles) Red Blood Cell (Erythrocyte) Model Molecule: Hemoglobin Heme Groups in Hemoglobin Hemoglobin – Tertiary Structure One beta subunit (8 alpha helices) Hemoglobin – Quaternary Structure Two alpha subunits and two beta subunits (141 AA per alpha, 146 AA per beta)