* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PDBe Motif
Protein (nutrient) wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Bottromycin wikipedia , lookup
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
Signal transduction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Biosynthesis wikipedia , lookup
Protein adsorption wikipedia , lookup
Homology modeling wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
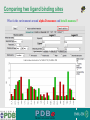
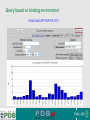
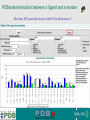
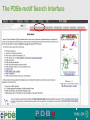
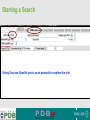
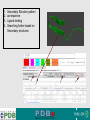
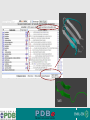
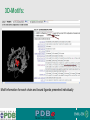
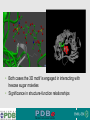
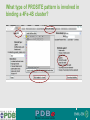
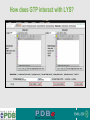
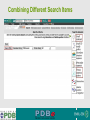
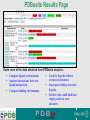
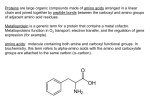
PDBemotif A web based integrated search service to understand ligand binding and secondary structure properties in macromolecular structures EBI is an Outstation of the European Molecular Biology Laboratory. PDBe-motif (http://www.ebi.ac.uk/pdbe-site/PDBeMotif) • Integrated search system for protein and nucleic acid structures. • User can construct a query based on a) Ligands and their 3D environment b) protein families (SCOP, CATH, UNIPROT, EC-number) c) protein secondary structures and different 3D motifs (PROSITE, beta turn, catalytic sites etc.) d) protein Φ/Ψ angle sequences • Results a) Sequence multiple alignment b) 3D multiple alignment of fragments, motifs and protein chains. c) Interactions statistics d) Motifs characteristics and properties distribution charts. 2 Comparing two ligand binding sites What is the environment around alpha-D-mannose and beta-D-mannose? Query based on binding environment What binds ASP ASP HIS LYS ? PDBesite-interaction between a ligand and a residue How does ATP generally interact with LYS in all structures ? The PDBe-motif Search Interface Different Search Options • PDB Header Search – PDB ID, Experiment Type, author name …. • Molecule Binding – Ligand 3-letter code, metal site geometry – looking for covalent, H-bonds, Van der Waals, planar interactions – distribution against amino acids, nucleic acids, ligands, Prosite motifs etc. • Pair bonds – interactions statistics between a pair of residues (Ligandaminoacid, ligand-ligand, ligand-nucleic acid etc.) • Motif binding – search for sequence pattern –distribute against ligands, secondary structures, 3D motifs • Search – combine different sources of data generate your query • Upload your own PDB file for analysis Starting a Search Using Sucrose Specific porin as an example to explore the site Results • Sequences • 3Dmotif • Ligand environment • Ligand bonds 1. 2. 3. 4. Secondary Structure pattern aa sequence Ligand binding Searching further based on Secondary structures 1a0t 3D-Motifs: Motif information for each chain and bound ligands presented individually Search Results for 1a0t –based on 3D motif configuration(195:IHWI,196:HWID) • Predominant secondary structure element – beta sheets Something Interesting!!!! 1a0t 1ofm • Predominantly beta sheet but tertiary structure quite different • Both cases the 3D motif is engaged in interacting with hexose sugar moieties • Significance in structure-function relationships Ligand Binding Environment • Residues are colour coded based on their nature of interactions with the ligand • Provides detailed 3dimensional information about the ligand binding environment Sucrose Binding • Aromatic amino acid sidechains (Tyr, Phe) are parallel to hexose/pentose sugar ring – helps in gliding mechanism • Lots of charged amino acids in the environmentfacilitating the transport Calcium Binding Lots of negatively charged amino acids What type of PROSITE pattern is involved in binding a 4Fe-4S cluster? Interaction Statistics •Clicking on each bar returns a list of PDB entries •The bars are proportionally colour coded based on the nature of interactions between the ligand and motif How does GTP interact with LYS? Statistics -Binding Interactions GTP vs Lys Colour coded based on nature of interactions Combining Different Search Items Pattern search Search for Small Molecule Combining both the queries Multiple 3D Alignment of GXXXXGKT motif/Walker A motif Particular amino acid sequence giving rise to secondary structure pattern – defining role in cellular processes PDBe-Site – Another Useful Tool to Analyse Ligand Environment Search options in the PDBe-site page • Define search by ligand • Define search by sequence motif (pattern) • Define search by metal site geometry • Define search by environment • has same environment • has similar environment Upload your own PDB file for analysis !! PDBesite Results Page Some uses of the data obtained from PDBesite analysis: • • • Compare ligand environments. Analyze interactions between ligand and protein. Compare binding environment. • • • Look for ligands within a certain environment. Superpose binding sites and ligands. Predict what could bind that empty pocket in your structure