* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Learning Classifier Systems - Operations, Information and Decisions
Nutriepigenomics wikipedia , lookup
Human genetic variation wikipedia , lookup
Genetic drift wikipedia , lookup
Public health genomics wikipedia , lookup
DNA paternity testing wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
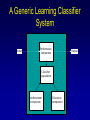
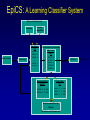
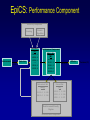
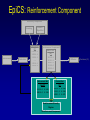
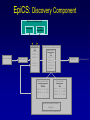
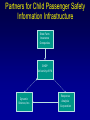
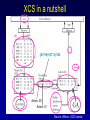
Evolutionary Computation Genetic Algorithms Genetic Programming Learning Classifier Systems Genetic Algorithms • Population-based technique for discovery of knowledge structures • Based on idea that evolution represents search for optimum solution set • Massively parallel The Vocabulary of GAs • Population – Set of individuals, each represented by one or more strings of characters • Chromosome – The string representing an individual The vocabulary of GAs, contd. • Gene – The basic informational unit on a chromosome • Allele – The value of a specific gene • Locus – The ordinal place on a chromosome where a specific gene is found Thus... Chromosome 011010 Gene (Allele="0") Locus=5 Genetic operators • Reproduction – Increase representations of strong individuals • Crossover – Explore the search space • Mutation – Recapture “lost” genes due to crossover Genetic operators illustrated... Parent 1: Parent 2: Parent 1: Parent 2: Parent 1: Parent 2: 011010 000110 011010 000110 011010 000110 Simple reproduction Offspring 1: Offspring 2: Reproduction with crossover at locus 3 Simple reproduction with mutation at locus 3 for offspring 1 011010 000110 Offspring 1: Offspring 2: 011110 000010 Offspring 1: 010010 000110 Offspring 2: GAs rely on the concept of “fitness” • Ability of an individual to survive into the next generation • “Survival of the fittest” • Usually calculated in terms of an objective fitness function – Maximization – Minimization – Other functions Genetic Programming • Based on adaptation and evolution • Structures undergoing adaptation are computer programs of varying size and shape • Computer programs are genetically “bred” over time The Learning Classifier System • Rule-based knowledge discovery and concept learning tool • Operates by means of evaluation, credit assignment, and discovery applied to a population of “chromosomes” (rules) each with a corresponding “phenotype” (outcome) Components of a Learning Classifier System • Performance – Provides interaction between environment and rule base – Performs matching function • Reinforcement – Rewards accurate classifiers – Punishes inaccurate classifiers • Discovery – Uses the genetic algorithm to search for plausible rules The Learning Classifier System • Rule-based knowledge discovery and concept learning tool • EpiCS – First Learning Classifier System designed for use in epidemiologic surveillance – Supervised learning environment Knowledge Representation • Classifiers – IF-THEN rules • Condition=“genotype” • Action=“phenotype” – Strength metric – Encoded as bit strings or numerics • Population – Fixed size collection of classifiers Low-level knowledge representation: The Classifier 0111*00011*111:0 Taxon 34.9 Action Bit Strength • Taxon is analogous to a condition (LHS) of an IF-THEN rule • Action bit is analogous to an action (RHS) of an IF-THEN rule • Strength is an internal fitness function High-level knowledge representation: Macrostate Population Number 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Classifier ****0**********:0 ****1**********:1 ****0*********0:0 ****1*********0:1 ****1********1*:1 ****0********1*:0 ****0*0********:0 ****1**********:0 ***00**********:0 0***0**********:0 ****0*****1****:0 *0**0**********:0 **1*1**********:1 **1*1*********0:1 ***00*********0:0 ****1**0*******:1 ****10*********:1 ****1*********0:0 Proportional Strength 0.39958000 0.38533600 0.08509200 0.08131500 0.02613600 0.00721500 0.00503500 0.00139700 0.00103500 0.00103400 0.00103100 0.00100800 0.00094900 0.00085500 0.00085000 0.00084300 0.00065600 0.00063200 N 380 382 94 91 28 8 5 2 1 1 1 1 1 1 1 1 1 1 Components of a learning classifier system • Performance – Provides interaction between environment and classifier population – Performs matching function • Reinforcement – Rewards accurate classifiers – Punishes inaccurate classifiers • Discovery – Uses the genetic algorithm to search for plausible knowledge structures Generic Machine Learning Model Input Performer Knowledge Base Learner Critic Output A Generic Learning Classifier System Input Performance component Output Classifier population Reinforcement component Discovery component EpiCS: A Learning Classifier System Discovery Component Covering Environment Detectors 10010 Genetic Algorithm Population [P] 01001:1 10010:0 1*010:1 --**110:1 1*001:0 Performance Component Match Set [M] 10**0:1 1**1*:1 1001*:1 ***10:0 100*0:0 Not-correct Set Not[C] ***10:0 100*0:0 0.560 0.334 1001*:1 Correct Set [C] 10**0:1 1**1*:1 1001*:1 Reinforcement/Penalty Regime Reinforcement Component 0.560 0.334 0.871 Effector Decision (=1) EpiCS: Performance Component Discovery Component Covering Environment Detectors 10010 Genetic Algorithm Population [P] 01001:1 10010:0 1*010:1 --**110:1 1*001:0 Performance Component Match Set [M] 10**0:1 1**1*:1 1001*:1 ***10:0 100*0:0 Not-correct Set Not[C] ***10:0 100*0:0 0.560 0.334 1001*:1 Correct Set [C] 10**0:1 1**1*:1 1001*:1 Reinforcement/Penalty Regime Reinforcement Component 0.560 0.334 0.871 Effector Decision (=1) Performance component • Creates a subset (the matchset, [M]) of all classifiers in population [P] whose conditions match a string received from the environment • From [M], a single classifier is selected, based on its strength as a proportion of the sum of all strengths in [M] • The action of this classifier is then used as the output of the system EpiCS: Reinforcement Component Discovery Component Covering Environment Detectors 10010 Genetic Algorithm Population [P] 01001:1 10010:0 1*010:1 --**110:1 1*001:0 Performance Component Match Set [M] 10**0:1 1**1*:1 1001*:1 ***10:0 100*0:0 Not-correct Set Not[C] ***10:0 100*0:0 0.560 0.334 1001*:1 Correct Set [C] 10**0:1 1**1*:1 1001*:1 Reinforcement/Penalty Regime Reinforcement Component 0.560 0.334 0.871 Effector Decision (=1) Reinforcement component • Correct set [C] is created from classifiers in [M] advocating correct decisions • Remaining classifiers in [M] form Not[C] • Tax is deducted from the strengths of all classifiers in [C] • Reward is added to the strengths of all classifiers in [C], biased for generality • Penalty is deducted from the strengths of all classifiers in Not[C] EpiCS: Discovery Component Discovery Component Covering Environment Detectors 10010 Genetic Algorithm Population [P] 01001:1 10010:0 1*010:1 --**110:1 1*001:0 Performance Component Match Set [M] 10**0:1 1**1*:1 1001*:1 ***10:0 100*0:0 Not-correct Set Not[C] ***10:0 100*0:0 0.560 0.334 1001*:1 Correct Set [C] 10**0:1 1**1*:1 1001*:1 Reinforcement/Penalty Regime Reinforcement Component 0.560 0.334 0.871 Effector Decision (=1) Discovery component • Genetic algorithm invoked once per iteration • One new offspring is created, from parents deterministically selected based on strength • The single offspring replaces weakest classifier in the population Features of EpiCS • • • • • Object-oriented implementation Stimulus-response architecture Payoff/Penalty reinforcement regime Syntactic control of overgeneralization Differential penalty control of undergeneralization • Ability to compute risk of outcome Discovering risk with EpiCS • Output decision of the learning classifier system is probability of disease (CSPD), rather than dichotomous decision • CSPD determined from proportion of classifiers matching a given input case’s taxon Discovering risk with EpiCS: The specifics Classifiers *10*0:1 0*100:0 01*00:1 0***0:1 CSPD Proportion in match set 0.60 0.05 0.22 0.13 (probabilities of classifiers associated with disease) (probabilities of all classifiers with matching taxa) 0.95 0.95 1.0 Discovery of Predictive Models in an Injury Surveillance Database: An Application of Data Mining in Clinical Research Partners for Child Passenger Safety Information Infrastructure State Farm Insurance Companies CHOP University of PA Dynamic Science, Inc. Response Analysis Corporation Why data mining is needed for PCPS • Large number of raw and derived variables renders traditional “manual” methods for discovering patters in data unwieldy • Hypothesis-driven (biased) analyses may lead to missed associations • Constantly changing patterns in prospective data require constantly changing analytic approaches that can be informed by data mining Candidate Predictors • Demographics • Kinematics • Characteristics of crash • Restraint use Outcome: Head Injury • Major burns involving the head • Skull fracture • Evidence of brain injury reported by respondent – Excessive sleepiness – Difficulty in arousing – Unresponsiveness – Amnesia after accident Data Preparation • Pool of 8,334 records • 20 separate datasets created – All cases of head injury included (N=415) – Equal number of non-head injury cases randomly drawn from pool • Each dataset randomly sampled to create mutually exclusive training and testing sets of equal size Comparison methods: Logistic Regression • Variables from training sets stepped into model to determine significant terms • Significant terms used to create new risk model: Pˆ y 1 1 e ( 1 x1 ... n xn ) • Risk model applied to cases in testing set • Risk estimates categorized by deciles and used construct ROC curves Comparison Methods: Decision Tree Induction • C4.5 used to create decision trees from training sets • 10-fold cross-validation used to optimize trees • Optimized trees used by C4.5RULES to classify cases in testing set Experimental Procedure for a=1 to maximum number of training epochs for x=1 to 100 Training present randomly selected training Phase case for x=1 to number of training cases evaluate training case x for x=1 to number of testing cases evaluate testing case x Genetic algorithm inactive Interim Evaluation Phase Training Epoch Trial Testing Epoch Results: Training 1 0.9 0.8 0.7 0.6 0.5 0.4 0.3 AUC 0.2 Indeterminant Rate 0.1 0 0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 Iterations Results: Training • EpiCS – 5,000 unique classifiers reduced to 2,314 by the end of training • Logistic regression – Single model with eight significant terms, no significant interactions • C4.5 – 11 rules created for each training set, most with single conjuncts Results: Prediction Area under the ROC curve obtained on testing, averaged over the 20 separate studies EpiCS 0.97 (.04) Logistic Regression 0.74 (0.03) C4.5 0.79 (0.04) And now for something a little different The XCS model XCS: A little history • Wilson, SW: Evolutionary Computation, 2(1), 1-18 (1994) – ZCS • Wilson, SW: Evolutionary Computation, 3(2), 149175 (1995) – The seminal work on XCS • Many papers by Lanzi, Barry, Butz, and others • Butz, M and Wilson, SW: Advances in Learning Classifier Systems. Third International Workshop (IWLCS-2000), Lecture Notes in Artificial Intelligence (LNAI-1996). Berlin: Springer-Verlag (2001) – The algorithm paper What is XCS? • An LCS that differs from traditional Holland model – Classifier fitness is based on the accuracy of the classifiers payoff prediction, rather than the prediction itself – The genetic algorithm is restricted to niches in the action set, rather than applied to the classifier population as a whole • The major feature is graceful, accurate generalization XCS in a nutshell ((43*99)+(27*3))/102 Action: 00 Action: 01 Source: Wilson, XCS tutorial EpiXCS: An XCS-Based Learning Classifier System for Epidemiologic Research Outline • What is it? • EpiXCS architecture – Data encoding – Evaluation metrics – Reinforcement – Missing values handling – Classifier ranking – Risk assessment • Test case: Pima Indians Diabetes Data What is EpiXCS? • Learning classifier system based on the XCS paradigm – Uses the Lanzi C++ kernel • Designed for use in epidemiologic research, specifically mining disease surveillance databases in supervised learning environments – Visualization by non-LCS users – Sensitive to demands of clinical data Data Encoding in EpiXCS • All numeric data formats permissible – – – – Binary Categorical Ordinal Real • Non-binary data represented using “centerspread” approach – Two genes per feature • Actions are limited to binary (for now) Sample input data format (Pima Indians Diabetes Database) ATTRIBUTE 0 <WILD "99"><REAL><STRING "Clump Thickness"> ATTRIBUTE 1 <WILD "99"><REAL><STRING "Uniformity of Cell Size"> ATTRIBUTE 2 <WILD "99"><REAL><STRING "Uniformity of Cell Shape"> ATTRIBUTE 3 <WILD "99"><REAL><STRING "Marginal Adhesion"> ATTRIBUTE 4 <WILD "99"><REAL><STRING "Single Epithelial Cell Size"> ATTRIBUTE 5 <WILD "99"><REAL><STRING "Bare Nuclei"> ATTRIBUTE 6 <WILD "99"><REAL><STRING "Bland Chromatin"> ATTRIBUTE 7 <WILD "99"><REAL><STRING "Normal Nucleoli"> ATTRIBUTE 8 <WILD "99"><REAL><STRING "Mitoses"> ACTION 9 <STRING "Malignant"> 5 4 4 5 7 10 3 2 1 0 3111223110 8 10 10 8 7 10 9 7 1 1 … Classifier Population Initialization • Minima and maxima for each attribute determined automatically at start of run • Center values can be initialized by user – Mean – Median – Random value between spread • Spread values can be initialized by user – Standard deviation – Quantile Sample Macroclassifiers /5.5,5.5/107.5,51.5/64.0,21.0/#/316.0,160.0/16.55,16.55/#/#/:1 /3.0,3.0/#/91.5,30.5/#/#/#/#/26.0,5.0/:1 /2.5,2.5/119.5,63.5/#/#/#/#/#/#/:1 /2.5,2.5/107.5,51.5/64.0,21.0/#/#/#/#/49.0,28.0/:1 /2.5,2.5/#/66.0,22.0/#/317.5,301.5/#/1.0040,0.9260/49.0,28.0/:1 /2.5,2.5/107.5,51.5/64.0,21.0/#/#/#/1.0735,0.9955/#/:1 Evaluation Metrics • • • • • • Sensitivity Specificity Area under the ROC curve Predictive values Accuracy Learning rate A Fast Primer on Test Evaluation Test Positive Negative Gold Standard Positive Negative 95 72 5 28 95 Sensitivity (True Positive Rate) 0.95 100 28 Specificity (True Negative Rate) 0.28 100 95 Positive Predictive Value 0.57 167 28 Negative Predictive Value 0.85 33 Sensitivity • Prior probability of a test-positive • If it’s high, then one would want to use the test to diagnose (classify positive) • If a classifier’s Se is high, then that classifier should be more likely to be used in defining an Correct Set when a training case is known positive Specificity • Prior probability of a test-negative • If it’s high, then one would want to use the test to rule out (classify negative) • If a classifier’s Sp is high, then that classifier should be more likely to be used in defining an Correct Set when a training case is known negative The Predictive Values • Posterior probability of a test-positive or negative • If a PPV is high, then once one has the test result in hand, and it predicts positive, it would be considered to be accurate • If a NPV is high, then once one has the test result in hand, and it predicts positive, it would be considered to be accurate How these metrics are used in EpiXCS • To evaluate classification performance – Training • Se, Sp, AUC, Accuracy, and Indeterminate Rate are plotted every 100th iteration – Testing • Se, Sp, AUC, Accuracy, and Indeterminate Rate are obtained for the testing set AUCShoulder 1000 Shoulder How these metrics are used in EpiXCS • To evaluate learning AUCShoulder 1000 Shoulder – Shoulder is the iteration at which 95% of the maximum AUC obtained during training is first attained, and AUCShoulder is the AUC obtained at the shoulder and classification performance Reinforcement in EpiXCS • Done the usual way, but… • User can bias the reward depending on the class distribution – Give disproportionately less “negative” reward to False Negative classifiers in data with <50% positives (where the Se is low) – Give disproportionately less “negative” reward to False Positive classifiers in data with <50% negatives (where the Sp is low) Missing Values Handling during Covering • Four possible ways to cover missing data in a nonmatching input σ that needs to be covered – Wild-to-wild: • Missing attributes covered as #s – Random within range • Random value within the range for the attribute – Population average • Population average for the attribute – Population standard deviation • Random value within the standard deviation for the attribute Classifier Ranking • After training, classifiers ranked according to their predictive values – Classifiers predicting positive ranked by PPV – Classifiers predicting negative ranked by NPV • Classifier ranking used for rule visualization Risk Assessment • Based on risk assessment module used in EpiCS • Risk estimates determined on testing based on proportional prevalence in match sets for each testing case • Provides risk assessment analogous that obtained by logistic regression Test case: Pima Indians Diabetes Data • 768 cases – 268 positive, 500 negative • 8 attributes – – – – – – – – Gravidity Plasma glucose Diastolic blood pressure Skin-fold thickness Serum insulin Body mass index Pedigree function Class: Diabetes Yes/No Experimental procedure • Training and testing sets created – 134 positives/250 negatives in each • EpiXCS – Results averaged over 20 runs, 50,000 iterations each • See5 – Boosting at 10 trials – 10-fold crossvalidation • Logistic regression – Relaxed stepwise model built on training set and evaluated on testing set Rules: EpiXCS If Number of times pregnant is 7.0 ± 7.0 and plasma glucose concentration after 2 hours is 67.5 ± 11.5 and triceps skinfold thickness is 33.0 ± 26.0 and 2-hour serum insulin is 326.5 ± 66.5 and age is 48.0 ± 27.0 Then not diabetes If Number of times pregnant is 7.5 ± 7.5 and triceps skinfold thickness is 35.5 ± 24.5 and 2-hour serum insulin is 811.5 ± 34.5 and body mass index is 48.9 ± 6.1 and pedigree function is 0.97 ± 0.89 Then diabetes Rules: See5 Rule 9/1: (20.5, lift 1.8) pedigree <= 0.179 age <= 34 -> class 0 [0.955] Rule 9/2: (62.1/2.6, lift 1.8) plasmaglu <= 103 pedigree <= 0.787 -> class 0 [0.944] Rule 9/3: (9.3, lift 1.7) serumins <= 156 bmi <= 35.3 age > 34 age <= 37 -> class 0 [0.912] Rule 9/9: (12, lift 2.0) plasmaglu > 135 serumins <= 185 bmi > 33.7 pedigree <= 1.096 age > 37 -> class 1 [0.928] Rule 9/10: (37.1/2.5, lift 2.0) plasmaglu > 103 bmi > 35.3 pedigree <= 1.096 age > 34 -> class 1 [0.909] The logistic model Risk of diabetes= 1.34 + 0.19*Gravidity + 0.04*Post-prandial glucose + -0.01*Diastolic blood pressure + 0.01*Skinfold thickness + -0.01*Serum insulin + 0.05*Body mass index + 0.72*Pedigree function Classification accuracy on testing EpiXCS Se 0.75 (0.06) Sp 0.96 (0.04) PPV 0.82 (0.02) NPV 0.83 (0.03) AUC 0.86 (0.04) Learning 0.04 (0.001) rate 0.79 0.92 0.59 0.89 0.86 Logistic Regression 0.61 0.89 0.75 0.81 0.86 --- --- See5 Conclusions • EpiXCS incorporates features of EpiCS into the XCS paradigm • Facilitates analysis of epidemiologic data • Uses metrics understood by clinical researchers • Discovers knowledge comparably to See5 and logistic regression