* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Clustering Method for Repeat Analysis in DNA sequences
Whole genome sequencing wikipedia , lookup
Molecular ecology wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Multilocus sequence typing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genomic library wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Copy-number variation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
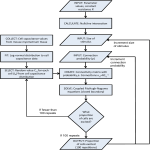
Clustering Method for Repeat Analysis in DNA sequences GNANA SUNDAR RAJENDIRAN JOYESH MISHRA RISHI MISHRA FALL 2008 BIOINFORMATICS Abstract Implement a Proposed Clustering Technique by Volfovsky et al. for Repeat Analysis Distribute Merging Repeats into Classes/Clusters based on Similarity Measures Why analyze repeats? Algorithm Description Selection using RepeatMatch Steps 1. 2. 3. 4. Preprocessing Merging and Repeat Map Generation Classification BLAST searches & further merging of Clusters Results RepeatMatch Uses Suffix Tree algorithm to determine all the exact repeats in a given sequence It is a part of the MUMmer package used for the rapid alignment of very large DNA and amino acid sequences. Example… Forward & Reverse Complement Repeats Definitions Exact Repeat: Subsequence that occurs in DNA at least twice. Exact repeat is represented by pair of co-ordinates (A1,A2) delimiting its location in the genome sequence and by the repeat length. Maximal Repeat: Repeat that can not be extended in either direction without incurring a mismatch Initial Repeat Set: Set of repeats chosen initially, from which the repeat classes will be constructed Definitions Preprocessing Output from RepeatMatch is used to partition the original genome sequence Each partition point has a reference to the pair coordinates (A1, A2) and repeat length l For each repeat starting at co-ordinates A1 and A2, with length l, this list will include both (A1, A2, l) and (A2, A1,l) Merging & Repeat Map Generation This procedure works by repeatedly working together two exact repeats that either overlap or that occur within a limited distance(a gap) of each other. Significant subsequences of the new merging repeats appear at least twice in the genome sequence. Merging with Gap Merging with Overlap Merging with Gap Given 2 partition points: p1 = (A1, A2, lA) and p2 = (B1, B2, lB) [A1 < B1] Compute the distance between the non-overlapping repeats as d(p1,p2) = max(0, B1 - A1 - lA + 1) Given a maximum Gap size G > 0 Sequences corresponding to p1 and p2 are merged if d(p1, p2) < G Merging with Overlap Merges sequences which are partially identical Overlap of 2 sequences is denoted as: o(p1, p2) = max(o, A1 + lA – B1 + 1) for A1<B1 Given a minimum overlap proportion op, where 0 <= op <= 1, repeat points (A1, A2, lA) and (B1, B2, lB) are merged if at least one of the four repeats has overlap satisfying o(p1, p2) > op min(lA, lB) op : Interpreted as a fraction of the shorter of the 2 repeats. E.g. if op = 0.75, the two overlapped sequences are merged if the length of their overlap is at least 75% of the length of the shorter sequence. Output: Merging Repeat The new sequence is defined as merging repeat with starting position M = A1 and with length lM = max(A1 + lA, B1 + lB) – A1 A data structure stores with each merging repeat its start co-ordinate, the length(nM), and a list of references to itself and to other repeats. Classification If a merging repeat has at least one reference in common with one another, then they belong to the same class If a merging repeat has references that belong to multiple distinct classes, then those classes are combined into one. If a merging repeat contains no reference to an existing class, then the merging repeat forms a new class. BLAST searches and further merging To merge similar but non-exact repeats Search all merging repeats against all others A local BLAST database is created with all repeat sequences using formatdb Classes are merged if any of their underlying sequences have a BLAST E-value less than a UserSpecified Threshold when compared to any sequence in another class If a class appears in multiple similarity pairs, all these similar classes are merged with the original class Results Results (contd.) Results (contd.) Results (contd.) Results (contd.) Results (contd.) Results (contd.) What’s Next? The results collected help to cluster similar repeats and generate a database. This DB, in future, can be used to find Similar Repeats faster. Also, if on future classification, we find common traits among repeats, they can be analyzed only for that cluster alone. Resources Softwares Used: RepeatMatch (MUMmer 3.0) BLAST (formatdb, blastall) MS Excel 2007 Programmming Language Used: C++, UNIX Shell Script Source for Project & Presentation www.cise.ufl.edu/~jmishra/bioinformatics.html References Volfovsky N., Haas Brian J. and Salzberg Steven L. A clustering method for repeat analysis in DNA sequences, 2001 Mummer software : http://mummer.sourceforge.net/ http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/ blastall, formatdb www.ncbi.nlm.nih.gov/BLAST/ References http://www.tigr.org http://www.animalgenome.org/blast/docs/blast_faq. html Of course, Wikipedia … Data obtained from: ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ Thank You