* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Analysis of the Brassica oleracea genome by the generation of B
Survey
Document related concepts
Human genome wikipedia , lookup
Pathogenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Koinophilia wikipedia , lookup
Genome (book) wikipedia , lookup
Y chromosome wikipedia , lookup
Genomic library wikipedia , lookup
X-inactivation wikipedia , lookup
Neocentromere wikipedia , lookup
Genetically modified organism containment and escape wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Transcript
Theor Appl Genet (1987) 74:758-766
D Springer-Verlag 1987
Analysis of the Brassica oleracea genome by the generation
of B. campestris-oleracea chromosome addition lines:
characterization by isozymes and rDNA genes
C. F.Quiros, O.Ochoa, S. F. Kianian and D. Douches
Department of Vegetable Crops, Umversily ofCalifomia. DavIs. CA 95616, USA
Received April 27, 1987: Accepted June 16, 1987
Communicated by C. S. Khush
Summary. This study aimed at generating chromosome
<.tddltion hnes and disclosing genome specific markers in
8m.15I('(1. These stocks will be used 10 study genome
evolutJvn in Bmss/('Ci ()ler(lCCo L. B, ('ampel/r!s Land
the derived am phidi plOJd ,,,,,eCles B. napus L B. cam
pes/f1S-o!f/'aceo mono,om ic and dlsomic cli "omosome
addition plants w.:re generated by crossing <lnd
backcrosslng the natural amphidiploid B. napLl!> 10 lhe
Jiplold parental species B, campeSlns. The pollen VI'
abtllty of the derived sesq uidiploid and hyperploid
ranged from 63S, to l:I8~; . whIle the monosomic ;lnd di
somic addition plants had an average pollen fCrlility of
94% and 9) %, respectively. The addition lines were
genetically chari;lcteri/\'d by genome specific markers.
The isozymes for 6PGD, LAP, PGI and PGM, and
rONA Eel! RJ reslnClton fragments were found 10 pos
seSS lhe desired genome specificity. Duplicated loci for
,,~veral of these markers were observed in B. campeslris
and R. o!UGCl'iI. <;upporling the hypothesis that these
diplOid ,penes are actually secondary polyploids. A to
lal of elghl monosom:c and eight disomlc addition
rlants were identi fil'd and characterized on the basis of
these markers. Another 51 plants remained un
characterized due to the lack of addItional markers.
rDNA genes were found to be distributed in more than
one chr"mosome, diffl'ling in its restriction sites. In·
tergenomlc recombination for some of the markers was
detected at II eq uencies between 6~c. and 20',", revealing
lhe feasibility of llllergenomic gene transfer.
Key words; Bml.>IC(/
nome - M 1rk..:rs
-
Cok crops - Cytogenetics - Ge
Jntrnduction
llle genus Bra.wca has been the subject of numer<)uS
cytogenetJc 'itudies (Prakash and Hinata 1980). The
large num ber of diploid species with t'L'oomic J) urn bers
rangi ng from x. = 7 to x = 12. and derived am phidipJoids
in lhe genus. mak.es it an attractive rese,Hch subject
(Mlzushima 1980). Most of the cytogenic research in
Bmss/c(I J1HS cen tered around the eI uCldation of the
origm \)1' the culllvatcJ am phldi plaids. B. "opus L. B.
carinll/a A. Be. and B. prnceo L. by Karpcchenko (1922),
Morin"ba (1934). and U (1935). This early work was
based on th..: synthesis of amphidiplolds by hybridi·
zation or the three basic dIploid cuhiva led species B. ni
gra L. (x = 8, genome b). B. oferocea L. (X" 9. genome c)
and B. campevlris L. (x = 10. genome a). A result of this
research was lhe poslUlaled tflangle of C, which IS a
diagrammatic representation of dIploid and amphi
dip 10ld species relationshIp (U 1935). More recently re
search by Robbins and Vaugha n (1983) on Rubisco and
by Palmer et al. (1983) <l nd Erickson et al. (1983) on
chloroplast DNA not only confirmed the validity of the
triangle of l' bu t also resolved lhe direclion of the
crosses leading 10 two orlhe amphidiploids.
Very ll\lIe IS known about the evolution of the diplQld
species and the origin of the different genomIc numbef$. On
lhe basis of chromosome painng in haploids (Keller and Arm
,trong 1983). di-gcllomic and {ri·genomic hybrids (Mlzushlma
1980; Prakash and Hinala 1980; Catcheside 1934. 1937; Sikh
1940). and l';'chy lene ka ryotypes (Robbclen 1960). II IS as
sumed lhal lhe baSIC genome of Brass/co is x = 6. Since no
,pecies Wllh 11115 genomic number has ever been reponed. they
are presumed [0 be exllnct. Robbelen (1960) found SIX basIC
lypes or chromosomes. designating them with the lellers A to
F, on [he baSIS of helerochromallC knobs and centromere posi
tion. He proposed Iha1 lhe composilion or each genome in the
culhvated dIploids was genome
a = AABCDDEFrr. b = AECDDEU and c'" ABBCCDEEF,
759
Two chromosomcs in each genome. namely A and C m B. nI
gra and B. olerclcea, and A in B. caml'eslris. were found 10 be
involved in the organizalion of the nucleolus. Thus, the diplOId
species are considered to be secondary polyploids, ,mce pre
sumably they have some of the baSK: chromosome lypes form
ing part of their genome in duplicale or even m Irlplicate. The
fact that chromosomes or the same lype wlthm or belween
species are not identical. indicales lhal rearrangemcnl, have
taken place during the evolution of the diplOid specIes.
In order to test these hypotheses based moslly on cytolo,2i
cal observations. cytogenetic slocks usd'ul to dissect each ge
nome and genome specific chromoso'tle markers are reqoired.
This approach has been very userul in Trilitum and relaled
genera (Hart and Tuleen [983), and Allium (Pelney el aL
19l>5), Genome specific isozyme lOCI In the Cultivated Brass/en
specIes have already been described (Cou;t!lan and Denford
1982: Aru sand Onon 1983. Arus 1984: Qui ros et a I, 1985).
In th is "udy. we rerNt the use Q( isozyme markers
Jnu rDNA genes III 1:1;,; charactuiza lion of B, cam
fle(lris-o/eracea l11onosom IL' and disomic addition lines
obla ined by crossl1lg and back crossing B, napll.\ 10 B.
wmpeslris, TIlese lin es wdl permit studies of genome or
,':aIlJzation and cvolu ti\Jo in Brassica.
1V13teri31s and methods
Plonl/?la/eria!
The lollowing acceSSIons were used: B napu:,. rapid cycling
CrGC -05 and rapid cycling cytoplasmic male Slenle CrGC-14:
R call1peslri.l, rapid cycling CrGe -0 I and rapId cychng cyto
plasmic mal.. staile CrGC-I3: rapeseed cv "Torch': Chinese
cabb~,~e cV 'K wan-Hoo Cho,' The rap,d cycling lines were ob
tainnl lJ'om Dr. Paul William, al the l.'niversity of Wisconsin.
USA (Williams and Hill 1986) Also. the lurnip cv 'White Lady'
was used to ,( ud I' the Hl hen ta nce of the triose phospha Ie
isom erase JSozy me,.
De' "iopmenl o/addfllon {me"
111e amphldlplold species B nupus was crossed eilher as pis
tillate or poJleo parent 10 the diploid B, campeSlrls. Embryos
were n:scued from developing 0' "les about 15 days afler pollJ
nJIIOn and cultured io sterile medium (Nitsch and Nitsch
1969). The resultmg sesqutdlpJold hybrids were backcrossed (0
B. (ampeSlfl5. Resulting progeny with more lhan 2n = 21 chro
mosomes were b~ckcrossed one or (wo more times to B. com
peSl flS. Mooosom ic addi tion lines. 20 = 2 J. were selfed to ob
la I n d ison11 c udJ it,on Ii nes (2 n = 22) Amon 8- all the crosses a
loul 0f ,',1)out 350 planls were generaled, The majorilY or these
were S! udied cy lologlcall y and eIeClrophoretically.
Chro(tlo\'ome ('(lunls
Flower buds were tixed [n propionic aeid: absolute ethanol
'iI,tll ferric chlond" added as a mordant (Swam lila than
ct 3.1 1954) After 24 h the huds were rinsed and SlOred 111 70%
tthanol Anlh<:,~ were d;~,ecled and squa~hed in a drop of I',.
a<:etoeannme for chromosome COllnl~. 10 to 30 cells were
exammed. PoJl;'n vlat>'''ly was determined on the basis of pol
len stamabllity In l't acetoCo'T111lne, A ml1l1mum of 100 pollen
grains was used for (his determination,
(J : 3)
ISI):!
me mark"r)
Horizon l(1I starch ge I el ecl [<J ph oreSI,' was u~ed to ~epara te the
enzymes obtained from a crude extracl of y()ung leave,.., and
poJJen leachales (Weeden and GOlli,cb 1980), Details of 11m
techmque have bt'c'n previously descnbed (Qujro~ and McHale
1985). TIle follOWing enzymes were assayed. 6phosphoglueo
nase deh yd rogen ase (6 PG D). ph 0sphogl uCOl$.Omerase (PG').
leuc ine UIn Inopeptidase (LA P). an d phosphogl ucom ula se
(PGM). Thc II1herllanee for lhe enzyme SVSlems LAP, PGI and
PGM hJve been reported In B. o!erGl:e; by Arus and Orlon
(1983) Although we did nol carry oul a formal genetic analysis
In R C(llJlpCS I fI.\. the SlmllanlY between lhe zymo:;rams of both
speCies mdlcak that they haw eq uivalenl loci c'oumg for lhese
cnzyme~.
rDNA ;;en('s
Total genomic DNA was lsol.ued from !eaves of indiVidual
plaots according to (he protocol of Saghal-Maroof et al. (1984)
wjlh the following modlficallons: the leaf lissue was h'lIr,)
gcnized With dry Ice in a coffee mIll (Mollhnex/Regal) (l<Jn<.lry
and Michelmore 1985) After extraction. the DNA was dlgesled
for 8 h with the endonuclease ErG Rl according 10 lhe manu
raerUl," tBRl). The DNA fragments were separaled by h<Jri
zoo tal ag" rose electro phoresls and transrered by Sou the rn blol
ling to Zeta-Probe membranes (Maniatis el al. 1982) Cloned
DNA from wheat rONA, probes pTA71 (Saghal- Maroof et al.
1984) and pTA250-2 (Appels and Dvorak 1982) was nick tranS
luted u.\tng standard lech mques (Mamatls el al. 1982) and
hybfldiLcd 10 til, membranes. D!'-.\ fragments were Sized u~
ing l:lmbd3 DNA as a refe.ence Radish (Raphanus SG/Il'US L.)
DNA was I~,ed as a control because lhe rONA genes of lhls
close relalJve of Brassica are well characlem:ed (Oelseny et al.
1983),
Results
SeSlluidlploid
hvtmd~'
111 f cross B. l1apus X B, campeslris and its reciprocal re
,;ulted in seeds devoid of endosperm, A tOIJI of seven
hyhrids were obtai ned after rescui ng and culturing em
bryos con (a Ined in these seeds. Four of the hybridS were
male-sterile due to the use of Ilk cy(oplasmlc male-<;tcr
ile ~locks as pistillate parents. The absence of fertility re
<;torer~ in the pollen parents made impractical the use of
(hese hy hrids in the development of the addition lines.
Therefore, our efforts were concentrated In the partially
!CrtiJc hybrid. 85 B138, wi th 65 % pollen viability (Table
I). A second partially fertile hybrid, 85 B 137, derived
Table 1. Chromosome numbers and
Brassil'O napus x B. campesrris hybrid.,
pollen
Viability
Plant no.
20
% Pollen viability
85869-1
85666-2
85676-1
85B146-1
85B137-1
85B 138-1
85665-1 '
30
15
ems'
ems
ems
67
65
ems
b
29
62
29
29
29
29
Reci rroca I cross
Cytopiasmic male Slerile
or
760
Table 2. Frequency of chromosome numbers and average pollen viability in plants derived from crossmg various hyperpJoid plants
to B. campesrris
0
I
0
0
0
0
0
0
1 (34)
1(19)
0
I
0
2
6
3
0
2
5
7
I
I
3
2 (30)
0
J (40)
2
2
96.3
94.2
91,2
87.6
85,0
83.1
83.3
3
7
2
2
0
0
20
21
22
23
24
25
26
Other'
1
8
6
I
26x20
J
% Pollen
viability
23x20
29x20
3
6
22x20
21 x20
24x20
2n
3
JO
3
0
0
0
5
(30)
• Chromosome nllmbers in parentheses
from the same cro,s, had the same level of pollen vi
a bili ly but Jl died shorlly afler Dowering.
Five of the seven hybrids had the expected chromo
some number of2n=29, whereas hybrids 85869-1 and
85 B76-1 had 2n = 30 and 2n = 62 chromosomes, respec
tively. 'llle last two perhaps derived by aneuploid
gametes. iollowed by chromosome doubling for 85B76
l. In general, the resultin~ hybrids morphologically re
sembled the B. campes/ris parenL characterized by slen
der leaves and spJOdly stems.
Table 3. Chromosom e numbers in pooled progenies of 2n = 21
and 2n = 22 B. campes/ris-o!eracea addition Lines
HyperplOid def! VOI/VCS
• No. of families
After back crossin" the sesq uidiploid hybrid 85 B138 to
L campes/TIS (2n = 29 x 2n = 20) 12 plants were obtain
ed. Two different strains of the la Iter were used. the rap
id cycling accession. CRGe-OI ane: the rapeseed cv
.OJ vrch', llle hybrid was used as Ihe pistillate parent
since the reciprocal cross resulted in poor seed set. The
seeds developed normally. with a full endosperm, mak
ing unnecessary the use of embryo culture. The plants
derived from '.he rapid cychng B. campeslris strain
nowcred q'.llte carly and ceased development shortly af
ler Dowering, In viL'W of this problem, we started using
Torch' as an alternative pollinator in some of the
crosses.
The pollen vlabilllY of the 14 plants derived from the
~ n = 29 x 2n = 20 crosses was 11 :gher than in the ses
qUllllploid; it ranged from 64% to 96%, with an average
of 81.1%. The avera 2c chromosome number of the hy
perplOld plants was 23,7, ranging from 2n=26 to
~ ) = 21, As the number of ch romosomes increased, the
pollen viability III these plants decreased. Thus. from
the first backcross to the diploid parent it was possible
to obtain at least one monosomic addition line (Table
2, In generaL these plan ts very much resem bled their
B. campeslris paren l.
The plants resulting from the 29 x 20 progeny were
crossed to B. Cilmpeslris. Chinese cabbage cv 'Kwan
, 100 Choi' in order to maximize leaf tissue and /lower
bud productIOn for the biochemical and cytological de
terminations, From these crosses. a number of plants
wi th 2n = 21 were obta ined. The average pollen viability
of these was 94,2 W The frequency of chromosome num
bers in the popled progenies of these plants is shown in
Table 3. Upon selfing. they yielded 2n=21 (25%) and
2n = 22 plants (19%), presumably monosomic and di
somic addition Jines. respectively (Fig. I a. b). The rest
were diploids. The B. a/eroceo extra chromosomes could
not be distinguished from the B. compeslris chromo
somes by the acetocarmine technique. All the 2n = 22
plants derived either from 2n = 21 plants or from higher
hyperploids were tentallvely dassi fied as either double
trisomtcs or as disomic addition lines on the basis of
chromosome pairing during diakinesis and metaphase I,
The puta tive disomic lines displayed II Jl in 60% to 80%
of thc cells, while the double trisomies displayed
1011 + 21 for most of the cells. Upan crossing the 2n = 2 I
plants to B. compes/TIS (2n = 20), about 30% of the
plants in the resulting progenies had extra chromo
somes. Selfing of double trisomic plants (2n = 22) or
crossing them to dJplOJd B. compeslris plants yielded
around 80% of plants with extra chromosomes. Selfing
of the lalter. however, resulted in twice as many pJant~
with 2n = 22 ch romosomes th,m selfing 2n = 21 plants
(Table 3). Only the progeny of a single disomic addition
line lor 6PG 0-2 was investigated. Upon seJfing or cross
ing to a diploid B. compes/ris plant, it yielded aboul
Progeny
n'
Frequency of chromosome
nos. (%)
20
2,,=21X2n=20
4
2n=210
2n=22x2n=20
2n = 220
4
4
3
21
6 (21)
8 (50) 4 (25)
6 (20) 13 (65)
9 (26) 3 (37)
19 (66)
22
Olher
3 (10)
3 (19)
4 (15)
4 (37)
I (3)
1(6)
0(0)
0(0)
761
50% and 30% of plants with extra chromosomes, respec
tively. About 50% of these har' ?n = 22 chromosomes.
Genome specific markers
The four enzyme systems listed in the "Materials and
methods" section were satisfactory [or identifying the
extra B. oferacea chromosomes present in the hy
perploid plants. Other system~ were tried. but the over
lapping or the complexity of the zymogTams precluded
their use as reliable markers. The best diagnostic en
zyme system was 6PGD. since it was found to be mono
morphic for a number of accessions of B. o/eracea, B.
CGlJlpeslris and B. napus (Fig. 2 a). This monomorphism
has meant that formal genetic tests have not been pos
sible for the determination of the number of loci in
volved in the syntheSIS of these isozymes. The
lY010grams of the diploids Showed clearly two activity
rones, the more anodal one, named 6PGD-1. is composed
of three equidistant bands. of which the most anodal
band is the only one shared by both species (Fig. 1 a, b).
For the more cathodal zone, named 6PGD-2. most B.
oferar:'a accessions display a three banded phenotype.
whd;; B compeslris has only one band overlapping with
the B. o/eracea band of slowest migration. Only the
6PGI)-1 isozymes persIsted in pollen leachates in both
diplOids, indicating cytosolic location. Furthermore the
lhro-'l: banded pa lIern observed In the leaves of B.
o/eroceo was also retaliled in pollen leachates revealing
durhcated loci for 6PGD-2 (Weeden and Gottlieb
1980). Conversely. the 6PGD-ll.;ozymes did not persist
In pollen leachates indicating plastid location. The
Isozymes from both diploid species were accounted for
In B. napus. confirming the hybrid nature of this amphi
diploid species. The multiple banded phenotypes for
6 PGD-l and 6 PG 0-2 bred true in selfed or sib pro
gen ies for each of the three species confirming the exist
ence of d uphcated loci.
Duplicated loci We)~ also observed for the genes
coding for tbe enzyme triose phosphate isomerase (Tpi
2 and Tpi-2') In the two diploid cultivated species. B.
oferacea and B. C(I111pCSIris. non segregating multiple
banded phenotypes were observed in leaves and in pol
kn leachates.. h us indica ting cytosohc location. On the
other h[lIiJ. the isozymes of the more anodal zone, TP1
I were Jo(;a ted in p!;t,tlds. followlOg the criteria of
Weeden and Gotlheb (1980). B. campeslris plants of the
\;lrnip cv 'White Lady' heterozygous for ooe of the
duplicat~d loci permitted the observalion of intergcnic
heterodimers forming between the monomorphic locus
and the two alleles of it~ polymorphic duplicate locus
(Fig. 2c). A sib cross between two TPI-2"/TPI-2'2 het
erozygotes segregated in the expected I: 2: 1 ratio,
10(2"): 20(2"12'2): 9(2'2). X2 = 0.03, P=0.99. For PGM
and LAP. isozymes migrating closer to the calhode and
common in B. oleracea were diagnostic for extra B.
o/eracca chromosomes (Fig. 3 c).
rDNA genes were fOund (0 be extremely useful for
the characterization of the hyperplOId hoes. Each dip
loid species has a lypical restriction pa llern with Eco RI,
although a few fragments in common were a:so ob
served. The restriction pa ltern of B. napus had all the
fragmlnts found in both diploid species (Fig. 3a). lhus.
Ihe 6PGD and the rONA systems were very reliable for
confirming the hybrid origin of B. napus. The res\r:ctlon
pattern of the radish used as control W;LS a combinalion
of those of B. oferacea and B. campeslris suggesting a
close relatIOnship between the two genera. In B.
o/eracea. the following fragments were observed: 6.0.
3.4. 1.8. 1.6, 1.4 and 0.8 Kb. while B. campeSlfIS dis
played 5.3, 4.1. 2.6. 1.6 and 1.4 Kb fragments. Radish
had a pallern similar to B. o/erucea, except for a 6.0 Kb
fragment whtch IS replaced by the two 5.3 and 4.1 Kb
fragments of B. w111peslris. All these flagments lighted
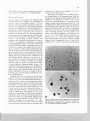
Fig. 1a, b. Pollen mother cells. a Mew.phase 11 for monosomic
addition line (2n = 21); 10 chromosomes al the leli and II al
the righl pole. b Metaphase I for a monosomic addition line
(2n '" 21). Showing a trivalent association
Fig.2. a 6PGD ~ymograms for
Brass/ca oleracea. B. comp"Slr;:, and
B. napus B. oleracea and B cam
pO'lm specific isozymes for 6PGD-1
and 6PGD-2 arc presenl lD lhe
hybrid specIes B. napur. b Inlerpre
tatIVe diagram for 6PGD-1. 6pgd-1
and 6pgd-1' are duplicated JOCL
Jsozymes J01 (B. oleracea) and I<:p
(B. camper/ris) at 6pgd-J have lhe
SJ me migr31ions. Isozymes 201 and
2cp at 6pgd-r form helerodimers
with lhe 6pgd.1 isozymes. T!lC
amphidiploid B. Ilapus (second line
from righl) and B. compeSlriS
oleracea addition lines (jar righl) diS
play an additIOnal hcterodimer
formed by the 2cp and 201 poly
peptides. The inlenslty of 20 I de
pends on lhe number of eXira
6PGD. B. oleracea chromosomes. c
Duplicated loci for the enzyme 11'[,
Tpi-2 monomorphic for lhe Isozyme
I and Tp/'}' polymorphic for the al
IOlymes 1 and 2. Firsr 16 lillt's from
lef! show segregating progeny
(1:2: I) for Tpi-1 allozymes I and 2.
Heterozygous individuals for Tpr.2'
(for example. line 2} form two
helerodimers, one belween the Tpi-2
isozyme and the Tpi-2' allozyme I
and another between the aJ)ozymes
I and 2 ofTpi-2'. Homozygous-indi.
vlduals for Tpi.2' for a single
helerodimer wilh Isozyme I of TpI-].
S PGO 1
f. f'G02
3pgd 1
101
1 cp
2cp
Il Pgd}'
2cp/201
(I.e. line J, TPI·2". line 7, TPI-2' L).
Lines 17 to the end show the pro
geny of a double homozygous plant
or phenolype TPl-]! and TPI-?'!
breeding true. Ine band at the
middle is lhe mterlOCIlS helerodlmer
Table 4. Distribution of B. oleracea-specilic alleles in B. campes/ris-o!eracea hyperploid plants and transmission 10 their progenies
Plant no.
2n
85B224-1
86B25-3
86B25-8
86B29-2
86829-6
85B224-3
86B44-1
85B268-1
116B55-3
86B56-2
851:l268-2
86850- i
86B170-3
86B170-6
86B50-2
86850-3
86850-7
86B 183-6
858268-4
86BI55·'
85B268-7
86B I09-2
24
21
PGI-I
6PGD-l
6PGD-2
LAP-I
~
~
PGM-2
RA
22
22
21
24
22
~:
"
22
'e
"'
22
~
21
21
22
*
*
22
21
22
22
*
'"
~
"
26
21
26
22
~:
*
'"
"
"
~
*
"
RB
763
Kb
RD CP NP I
CP-OL
I NP OL
6.0
5.3
4.1
Table 5. Nun,r,er of monosomic and disomic addition plaoLS
generated from S, napus X E, campes!ris crosses
Marker
POI-I
LAP-I
3·4
2 6
1.8
1.6
1.4
6POD-1
6FGD-2/RA
PGM-2
Unmarked
Monosomic
I
I
1
5
0
32
Disomic
I
2
I
4
1
19
the autoradiograms after hybridization with the
pr",be pTAljO,2 which carried only transcribed se
10
0.8
quence~.
Gene/it c!wrac/erizmion of hyperploid plants
l'
2
Fig.3a-c. Characterization of the B. campes/ris-oleracea ad
dition lines by chromosome markas. a rDNA EcoRI fragments
for radish (rd). S, campemis (cp), S, napus (np), four B. cam
pesrris-o/eracea hyperploid plants (2n ~ 23. 2n = 24. 2n -~ 26,
2n = 25) and B, o/cracea, Fragmenl~ of D. oleracea and B, cam
pes/r/S add up In the amphidiploid S, napus. Hyperploid plants
lack the B. o/eracea 3.4 Kb fragment. b B, campes!ris B,
oleracea mono omic addition lines Il,,. 6PGD-I' od 6PGD-2,
Arrows poinl 10 S, o/eraceo-specific lsozymes, c B. wmpestris
olera£'eG monosomic addition line for Pgm-2 (lines 7 and 8, ar
row points to D, a/eracea-specific isozyme), First two lines cor
respond to D. nopus; the rest show the D. C(Jmpestrls PGM
phenotype
Plants from all (he crosses and backcro~.ses described
above were ~ystematically surveyed for B. oleracea
isozymes, Pro,genies from hyperploid plants carrying B.
o!eroceo speciilc markers were screened for chromo
some number and for lhe presence of these markers.
Table 4 shows a sample of the phenotypes of some of
the hyperploid plants and the transmission of the B.
oleracea specific isozymes to 2n = 21 and 2n = 22 indio
viduals found in lheir progenie~. We were able to
clwradcriLL: eight 2n = 2 J plants as monosomic addition
lines, :\ild nine 2n "" 22 plants as disom ic addition lines,
by the presence of B. o/erocco specific alleles for the loci
sampled, II was found that 32 20 = 21 and 19 2n = 22
plants remained uncharaclerized due to the lack of ad
ditional m:t'rkers (Table S). For the rONA determi·
na()0n, we screencJ four hyperploid plants with 2n "". 23,
24,25 and 26 chromosomes, Two oflhem showed the si
multaneous presence of the B. o/Naceo 1.8 and 0 8 Kb
fraj!.ments in addition to the B. compeslris fragments in
dicating (he preseoce of a B. oleracea chromosome
car1)ing Ibese fragments, The rest had the B, campnlris
or B, lIapus rONA phenotypes. The presence of Ihese
two fragments was designated as RA phenotype
(Fig. 3a), 111e presence oC (he B. oleracea 3A Kb frag
ment was designated as RB phenotype, Thus, plants
carrying all three fragments had RA, RB phcnolype.
Three plants of20 = 21 and one oC2n= 22 chromosomes
derived Crom the hyperploid parents mentioned above
displayed the same phenotype as the parental plan IS,
The presence of lhe B. oleracea specific fragm cnts 1.8
and 0,8 Kb was accompanied by the S. oleracea 6PGD
2 isozymes, indicaling the genes coding lor these iso
zymes and the rONA genes are located on tltt: same
chromosome. On the other hand. the presence of lhe B,
olera<:ea 6PGD-l isozymes were independent from thai
oC the 6PGD-2 isozymes revealing that these loci are on
diJTerent chromosomes (Fig, 3 b). Similarly the presence
764
Fig. 4. Progeny from hyperplQid B. campeslris-o(eracea plant
carrying a PGI -2 8, oleracea-specific chromosome. Addition
lmes are heterozygous (3 banded phenotype) and diploid B,
('ampeslris are homozygous ror the tsozymes of raster migra
': Ill. The plant homozygous for lhe slower isozymc may have
originated by intergenomlC reoombinallon
Table 6. Frequency of intcrgenomlC recom billanLS observed in
the progenies of hyperplolds
Progeny
n·
85B 138-1
86B ISO-I
86B150-7
85B268-4
10
12
16
. Progeny SJze
5
Recombination
frequency (?OJ
20.0
10.0
8.3
6.2
Loci
Pgl-l
Pgi-/
6pgd-2
6pgd-f
of LAP-I. PGI-I and PGM-2 B. oleracea specific ISO
zymes were found to be Independent of each other.
Intergenomlc recombinants were observed in some
of (he progenies from hyperploid plants. A few diploid
plants showmg both B. o!eracea and B. campeslris iso
lvmes (Table 6) were delected. In addItIon, two plant.,
were homozygous for a B. oleracea POI Isozyme, lack
mg the B. wmpt:5lns isozymes (Flg.4). The sporadic
presence of multivalenls in some oC the addition lines
explains the origin of these r.:com binan LS (Fig, I b).
In progenIes from hyperploId plants we detected
eight self·compatible planls of either 2n = 21 or 22 chro
mosomes derived from crosses not involving the Chi
nese cabbage ·Kwan-Hoo Chai'. a cultivar with a very
relaxed self incompalJbility. [n addition. lWO diploid
plants derived from the same hyperploid parents were
also found to h,: self-compatible.
Discussion
Th e hIgh pollen viability in the sesq uidiploid hybrids
and In the sub equent hyperplOid derivatives permits
generation of a series of alien chromosome addItIon
lines in Bmssiw, A good example of the high tolerance
for exlra chromosomes In B. campesrris is shown by
2n = 26 plants (lJ,playing a p"lIen viability of 8]« TIle
tolerance for extra chromosomes might have evolved in
this species as a step to JJ(oploldization in the gener
ation of the hybrid polyploid B, napus. A similar situ
ation occurs in wheat where a whole series of aneuploid
stocks has been constructed (Sears 1969). The mO'lO
somic and disomic addition lines generaled in our study
had a pollen viability of at least 90%. Furthermore, the
transmission of the extra chromosome through Ihe
ovules of :' I] = 21 plants was on the average 21 %. lile
presence of disomic addition lines at a frequency of 10%
indicates that the extra chromosome undergoes non-dis
junction in Ihe female gametes, Selfing or monosomic
addition plants increased the transmission of the exll':)
chromosome resulling in 25% tri~omics and 19% tetra
somic plants In the progeny. This indicates that the ex
tra chromosome may be transmitted through pollen.
The 6PG1) loci and lhe rDNA phenotypes confirm lJ'e
Oflglll of B. 110pUS as a hybnd of B, ('ampes/ris and B. oleroao,
Allhaugh the number of loci duplicated for 6PGD could not be
pinpointed by genl'lic analysis du<: to the lack llf poly
morphism. they can be eXlrapolalCd by Jnspection of the
zymograms. Figure 2 b shows the interpretation for 6PG D I
based 00 2 loci designated 6pgd-1 and 6pgd-J' for each of the
diploid species, Locus 6pgd-1 seems to be monomorphIC for
both speCies, carrying t:\e allozymes lo( and lep or identical
nllgrallon, The duplicaled locus 6pgd-J' has genome speCIfiC
aUozymes 201 and 2cp for B. oleracea and B, campesirls. respec
tively. 111e middle bands ror each dIploid are in(c;locus
heterodimers. I n E. napus the allozymes 201 and 2cp f<11 m an
additional Illtcrlocu.,>, interspecific heterodlmer right below the
B. oleracea IOterlocus helerodilller. The same pallern .s ob
served in the monosomic addilion line~ for 6PGD-I. except
thal the allozyme 201ls weak due to the presence or only one
copy orlne B, oleroCf:o chromosome.
The distinct pattern of rONA fragmc:nts in R
uleracea and B, campeslris is useful for the detection
and characterization of the addition lines. The maio dif
ference i~ the replacement in B. compeslris of the B.
olerocea fragments 1.8 and 0,8 Kb by a 2.6 Kb fragmenl,
resulting from the loss of a reslriction site. The sekctive
loss of the rONA B, olerace(l 3.4 Kb fragment (RB
phenotype) and the simultaneous presence of the 1,8
and 0.8 Kb fragments in the addition lines indicates that
more than one chromosome carry these genes tn
Brass/c(l and that they have different restriction siles.
One chromosome carries the 1.8 and lhe 0.8 Kb frag
ments while other carries the 3,4 Kb fragment. This dlf
fermce In restriction sites indicates initial duplication
and subsequent divergence of these seq uenees. The
hg;l ter intemi ty of Ihe 1.8 and 0.8 Kb Cragments in lhe
addition lines is explained by the presence of a single
copy or the RA B, oleTl1cea chromosome versus two
coptes in B. /Jupt/s. In our limited SUn'ey for rONA
phenotype~ we did not detect any addi:ion lines of only
RB phenotype. Furlher testing of the unmarked addi
tton hnes i~ expected to disclose individuals with this
phenotype. The organization of the other fragments in
these two chromosomes is oot known at thIS point.
765
An important observation was the presence of pos
sible recombinants in some of the progenies, lnterge
nomic recombination has been reported between the B.
oferaceG and S, campeslris genomes by Chiang and
Crete (1983) after transferring a disease resistance gene
from B. IWPUS to B, olerGcea. In the amphidiploid no
evidence of recombination between the two genomes
has been reported. 1l1is lack of recombination is likely
due to the high diploidization of B. napus resulting in
J 911 in meiosis. After baekcrossing it 10 the diploid B.
CGl'llpeSlris parent. the loss of paiLng partners for some
of the chromosomes migh t result in an increased change
of multivalent formation and intergenomic recombi
naUon, A possibility that needs further exploration is
the prescilce or a paji I ng control mechanism sim ilar to
tiwt reported in wheat (Riley et al. 1959: Altia and Rob
he len I986). In any e\ en t, reco mbina tion open s th e pos
si bilit)' of exchanging genes among genomes, an im por
tant alternative for the lJrassica breeder. Recombination
between th~ B. a/ero( ea and B. campeslris genomes sup
ports the view that these species have angina ted from a
common ancestral genome by aneuploidy and chromo
some repattern ing. 'I hese changes migh t have been ex
pedited by translocations which not only rearrange the
chromosomes in novel comhinations but also yield ter
tIary IflSomies by dlstu rballee, in chromosome dis
junction (Stebbi ns In 1: Khus 1973).
The breakdown of self-mcompatlbllny In some of Ihe
derivative, is also an !I1lerestmg findmg. Self-compalibilily
could not b" c'«socI3ted with any speClfk B. oleracea marker. or
even With Ih", eXira chromosomes. since IWO diploids were
found to be compntible, 111e pOSSibility eXIsts. however. thal
haVing the S locus in a trisomic condition might weaken its
expressIOn, resulting in self.comrJtible plants, as occurs in the
natural amphIdiploid species which are selr-compatible. In
such a case, it might be possible to locate the S locus by using
these addition lines,
Our work opens the r>ossibihty of generating useful
cy logen dic stocks for tite genetic and cvolu tlOnary
chamCierizallOn of Br05S!cO specie.,. 1l1c presence of duplicated
locl ,n the B, (){eroceo and B. compeslris genomes agrees with
the hypNhds of Robbelen (1960), which suggests that the
bdS1C dLplolds are mdeed secondary polyploids. Olher evidence
is the rema rka ble tol erance of an eu plOldy and Ihe high fertility
of the aneuplOlds found m (Lis siudy, Generalion of addnion
lines for each of Ihe dIplOid genomes, Including x = 7 genomes
from several WIld speCie, IS underway. This WIll allow a
com raral;ve study of the Rrass/C(I genomes, as additional
markers are developed. A further step will be the generation of
synlhelle
amph,dlploids
between
diploids
carrying
agronom,eallv useful genes such as those determining disease
res I' ia nee, pia nl arch i!eCIll re and presence of importa nt
chemICal compounds, Generalion of addition lines from these
will provide information on the loeation of genes deterrnllling
til e va no us hOri'cu It IJ ral tra its observed in the diploids, i, e,
curd III cauliflower. headmg in cabbage, root enlargement in
turn! ps an d ax III 'Hy bud en largemen tin Brussels sprou ts.
kkno\<,'h'dgl:menrs. We are lIldepled to Charles Rick, Steve
ranksley, Judy Greenlee and Margi Oard for reviewing the
manuscript: to Vince D'Antonio. )anet Sutes and Mitch
McGru lh for tech meal assistance and to Jane Joh nson for
typlllg the manusenpt. Supported by a USDA wmpditive
granI86CRCR-I-1926.
References
Appels R, Dvorak J (1982) RelatJVe oaks of divergence of
spacer and gene sequences within thc rONA rLgion of
species in the Trillu:ae: implications for maintenance of
homogeneity of the repeat"'d gene family. Theor Appl
Genet 63: 361-365
Arus P (1984) B. oleroceo and B. nopus ,sozymes. Cruci fe rae
Newsktt 9:64
Arus P. Orton TJ (1983) Isozyme and linkage relationships of
Isozyme loc' in Brassica oleracea. J Hered 74:405--412
AllIa 1. Robbelen G (1986) Cytogenetic rehtionship withm
cultivated Brass/co analyzed in amphihaploids from three
diploid ancestors. Can j Genet CYlol 28: 323-329
Cateh eside DG (1934) The ch romosoma I relalionshlps 1Il the
swede and turnip groups of BraSSica. Ann Bot 48: 60 I 633
CJ.lcheSlde DO (1937) Secondary pairmg In Brass/co olaoceo
CytOIOglJ, Jubilee, pp 366-378
Chiang MS, Crete R (1983) Transfer of resistance 10 race 2 of
PlasmodlOphora brassicae from Brossica "opus to cabbage
(B. oleraceG ssp. COpi/OIG), V The inheritance of resiSlance.
Eu ph ytlca 32: 479-483
Coulthart M, Denford KE (1982) Isozyme studies in BrassiCG.
I. Electrophoretic techmques for leaf enzymes and corn
panson of B. napus. B, COmpl:SlflS and B. oleracea using
phQsphoglucom utase. Can J Plan t Sci 62: 621-630
Delseny M. Cooke R. Penon P (1983) Sequence helerogenelty
In radish nuclear ribosomal RNA genes. Plant SCI Lett
30: 107-109
Enckson LR, Siraus NA. Beve rsdorf WD (1983) ReslflctlOn
pallerns reveal onglns of chloroplast genomes In Brass/fG
amph id iploids, Theor Appl Ge net 65: 201- 206
!lart GE, Tu k~n NA {1983) Ch romosoma I location of ek ve n
E/,/lrigio efongala (= Agropyron elongalum) isozyme struc
tural genes, Genet Res 41: 181-202
KaflJechenkQ GD (1922) The number of chromosomes and the
genetic correlation of cultivated Cruciferae. Bull Appl Bot
Genet Plant Breed 13: 3-14
Keller Wk Armstrong KC (1983) Productton of haplOIds vIa
anlher culture in BroSS/ca oleraceo var ilahc(l, Euphyuca
32:151-159
Khush GS (1973) Genetic; Qf aneuploids, Academic Press,
New York, 301 pp
umdry BS. Michel more RW (1985) Selection of probes for re
striction fragment length analysis from plant genomic
clones. Plant Mol Bioi Rep 3: 174-179
Man,atlS T. Fritsch EF, Sam brook J (1982) Molecular cloning,
a laboratory manual. Cold Springs Harbor Laboratory,
Cold Springs Harbor, New York, 545 pp
Morinaga T (1934) On the chromosome number of Brassica
jUlIC/:a and S, nopus, on Ihe hybrid between thc two. and on
offspring line of the hybrid. Jpn J Genet 9' 161-163
Mlzu,hlma U (1980) Genome analysis in Bramca and allied
genera, fn: Tsunoda S, Hinata K. Gomez-Campos C (eds)
Bra,mco nops and wild all ie;, Japan SClen ti fie Sonelles
Press, Tokyo. pp 89-106
Nitsch JP, "LISch C (1969) Haploid plants from pollen grains,
Science 163: 85
Palmer J D, Shields CR, Cohen DB, Orton TJ {1983) Chloro
plast DNA evolution and the Origin of amphidiploid
BroIs/ca ;peCles. llleOf Aprl Genet 65: 18 1~ 189
766
Perney EB, Corgan IN, Horak KE, Tanbiey SD (InS) Elec
trophoretic analysis of AllIUm alien :"idJllon lJOes. Theor
Appl Genet 71: J 76- 184
Prakash S, Hinata K (1980) Taxonomy, cytogenetics and ongin
of crop Brassicas, a review, Opera Bot 55: I-57
Quiros CF, McHale (1985, Genellc analysIs of Isozyme vari
ants in diploid and tetraploid potatoes, Genetics
111:131-145
Quiros CF. Kianian SF. Ochoa 0, Douches 0 (/985) Genome
evolution in Brussicu: use of molecular markers and cy·
togenetic stocks, Cruci ferae Newsle II 10: 21-23
Riley K Chapman Y, Kimber G (1959) Genelle control of
chromosome pairing in intcrgcaeric hybnds With wheal.
Nature 183:1244-1245
Robbelen G (1960J Beitr1\ge zur Analyse des BraSSlca
Genomes. Chromosoma I): 205-228
Robbins MP, Vaughan 'JG (J 983) Rubisco in Brassicaceae, In:
Jensen U. Fairbrothcrs DE (cds) Proteins and nucleiC acids
In plant systematics. Spri'.'gcr. Berlin. pp 191-204
Saghai-Maroof MA. Sollman KM, Jorgensen RA Allard RW
(1984) Ribosomal DNA spacer-length polymorphisms In
barley, lvkndelian inherilanct'. chromosomal locallQn, and
population dynamiCS, PNAS lSA 81:80!4·-8018
Sear$ ER (1969) Wheal cytogenetics. Annu Rev Genet
3:451-468
Sikka SM (1940) Cytogenellcs of Bramca hybrids and species.
J Genet 40: 441 - 509
Stebbins GL (1971) ChromosQmal evolution III hIgher plants
Addison- Wesley, California
SW31lllnathan MS,' Magoon ML. Mehra KL (1954) A sImple
propionlC.carmine PMC smcar method for plants with
small chromosomes, Ind J G~r.et Plant Breed 14:87--88
U. N (1935) Genomic anaf"SIS JO Brassia} wtth spedal
reference to the expenmental formatIOn of B, napils and
peculiar mode QfferllhZ<l.lion.lpn J Genet 7:389-452
Weeden NF. Goltlieb LD (1980) Isolation of cytoplasmic en
zymes from pollen, Planl PhysiQI 66: 4DO-403
Williams PH, Hill CB (1986) Rapid-cychllf populations of
Brassiw, SCience 232: 1385-1389