* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Amino Acids
Fatty acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Genetic code wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Catalytic triad wikipedia , lookup
Metalloprotein wikipedia , lookup
Peptide synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Proteolysis wikipedia , lookup
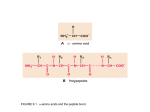
Chapter 1/Structure I • The Building Blocks • Chemical Properties of Polypeptide Chains Levels of Protein Structure • The AA sequence of a protein's polypeptide chain is called its primary structure. • Different regions of sequence form local regular secondary structures, (a-helices or -strands). • Tertiary structure is formed by packing structural elements into one or several compact globular units called domains. • The final protein may contain several polypeptide chains arranged in a quaternary structure. By formation of structures, amino acids far apart in the sequence can be brought closer together to form a functional region, called an active site. Amino Acids • • • • • • • Organic compounds with amino and carboxylate functional groups Each AA has unique side chain (R) attached to alpha (α) carbon Crystalline solids with high MP’s Highly-soluble in water Exist as dipolar, charged zwitterions (ionic form) Exist as either L- or D- enantiomers Almost without exception, biological organisms use only the L enantiomer Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011; Berg JM, Tymoczko JL, Stryer L, Biochemistry, 5th Edition, 2002 Formation of Polypeptides • Polypeptides and proteins are created through formation of peptide bonds between amino acids – Condensation reaction Peptide linkages R + H3N CH NH C CH O R O R C CH NH Polypeptide http://en.wikipedia.org/wiki/File:AminoacidCondensation.svg O C O - Polypeptide Chain • In a polypeptide chain the carboxyl group of the amino acid n has formed a peptide bond, C-N, to the amino group of the amino acid n + 1. One water molecule is eliminated in this process. The repeating units, which are called residues, are divided into main-chain atoms and side chains. The mainchain part, which is identical in all residues, contains a central Ca atom attached to an NH group, a C'=O group, and an H atom. The side chain R, which is different for different residues, is bound to the Ca atom. The “Handedness" of Amino Acids. • Looking down the H-Ca bond from the hydrogen atom, the Lform has CO, R, and N substituents from Ca going in a clockwise direction. For the L-form the groups read CORN in the clockwise direction. • All a.a. except Gly (R = H) have a chiral center • All a.a. incorporated into proteins by organisms are in the L-form. Amino Acids • 20 amino acids specified by the genetic code, grouped by different properties associated with R group (residues) http://courses.bio.indiana.edu/L104-Bonner/Sp11/imagesSp11/L12/Part1MPs.html Hydrophobic Amino Acids Charged Amino Acids Polar Amino Acids Chemical Structure of Gly • Glycine Gly G Glycine • Relative abundance 7.5 % • flexible, seen in turns Chemical Structure of Ala • Alanine Ala A Alanine • Relative abundance 9.0 % • hydrophobic, unreactive, a-helix former Chemical Structure of Val • Valine Val V Valine • Relative abundance 6.9 % • hydrophobic, unreactive, stiff, -substitution -sheet former Chemical Structure of Leu • Leucine Leu L Leucine • Relative abundance 7.5 % • hydrophobic, unreactive, a-helix, -sheet former Chemical Structure of Ile • Isoleucine Ile I Isoleucine • Relative abundance 4.6% • hydrophobic, unreactive, stiff, -substitution -sheet former Chemical Structure of Met • Methionene Met M Methionine • Relative abundance 1.7 % • thio-ether, un-branched nonpolar, ligand for Cu2+ binding a-helix former Chemical Structure of Cys • Cysteine Cys C Cysteine • pKa = 8.33 • Relative abundance 2.8 % • thiol, disulfide cross-links, nucleophile in proteases ligand for Zn2+ binding -sheet, -turn former Disulfide Bonds • Disulfide bonds form between side chains of two cysteine residues. • Two SH groups from cysteine residues, which may be in different parts of the AA sequence but adjacent in the 3D structure, can be oxidized to form one S-S (disulfide) group. • Usually occurs in extracellular proteins. 2 -CH2SH + 1/2 O2 -CH2-S-S-CH2 + H2O Chemical Structure of Pro • Proline Pro P Proline • Relative abundance 4.6 % • 2° amine, stiff, 20 % cis, slow isomerization seen in turns • Initiation of a-helix Chemical Structure of Phe • Phenylalanine Phe F Fenylalanine • Relative abundance 3.5 % • hydrophobic, unreactive, polarizable absorbance at 257 nm Chemical Structure of Trp • Tryptophan Trp W tWo rings • Relative abundance 1.1 % • largest hydrophobic, absorbance at 280 nm fluorescent ~340 nm, exhibits charge transfer Chemical Structure of Tyr • Tyrosine Tyr Y tYrosine • pKa = 10.13 • Relative abundance 3.5 % • aromatic, absorbance at 280 nm fluorescent at 303 nm • can be phosphorylated hydroxyl can be nitrated, iodinated, & acetylated Chemical Structure of Ser • Serine Ser S Serine • Relative abundance 7.1 % • hydroxyl, polar, Hbonding ability • nucleophile in serine proteases phosphorylation and glycosylation The Catalytic Triad of Trypsin Chemical Structure of Thr • Threonine Thr T Threonine • Relative abundance 6.0 % • hydroxyl, polar, Hbonding ability, stiff, -substitution phosphorylation and glycosylation Chemical Structure of Asp • Aspartic Acid Asp D AsparDic • pKa = 3.90 • Relative abundance 5.5 % • carboxylic acid, in active sites for cleavage of C-O bonds, member of catalytic triad in serine proteases, acts in general acid/base catalysis, ligand for Ca2+ binding Calcium-binding Site in Calmodulin Chemical Structure of Glu • carboxylic acid, • Glutamic Acid 2+ binding, ligand for Ca Glu acts as a general acid/base E in catalysis for lysozyme, proteinase GluEtamic • pKa = 4.07 • Relative abundance 6.2 % Chemical Structure of Asn • Asparagine Asn N AsparagiNe • Relative abundance 4.4 % • Polar, acts as both H-bond donor and acceptor molecular recognition site can be hydrolyzed to Asp Chemical Structure of Gln • Glutamine Gln Q Qutamine • Relative abundance 3.9% • Polar, acts as both H-bond donor and acceptor • molecular recognition site can be hydrolyzed to Asp N-terminal Gln can be cyclized Chemical Structure of Lys • Lysine Lys K Before L • pKa = 10.79 • Relative abundance 7.0 % • amine base, floppy, charge interacts with phosphate DNA/RNA forms schiff base with aldehydes (-N-N=CH-) • a catalytic residue in some enzymes Chemical Structure of Arg • Arginine Arg R aRginine • pKa = 12.48 • Relative abundance 4.7 % • Guanidine group, good charge coupled with acid charge interacts with phosphate DNA/RNA a catalytic residue in some enzymes Guanidine group Chemical Structure of His • imidazole acid or base; • Histidine pKa = pH (physiological), His member of catalytic triad in H serine proteases Histidine ligand for Zn2+ and Fe3+ binding • pKa = 6.04 • Relative abundance 2.1 % Distribution of Amino Acids • Codons (3 RNA bases in sequence) determine each amino acid that will build the protein expressed Properties of the Peptide Bond • Each peptide unit contains the Ca atom and the C'=O group of the residue n as well as the NH group and the Ca atom of the residue n + 1. • Each such unit is a planar, rigid group with known bond distances and bond angles. R1, R2, and R3 are the side chains attached to the Ca atoms that link the peptide units in the polypeptide chain. • The peptide group is planar because the additional electron pair of the C=O bond is delocalized over the peptide group such that rotation around the C-N bond is prevented by an energy barrier. Resonance Tautomers of a Peptide Peptide Bond • The peptide bonds are planer in proteins and almost always trans. • Trans isomers of the peptide bond are 4 kcal/mol more stable than cis isomers => • 0.1 % cis. Polypeptide Chain • Each peptide unit has two degrees of freedom; it can rotate around two bonds, its Ca-C' bond and its N-Ca bond. • The angle of rotation around the N-Ca bond is called phi (f) and that around the Ca-C' bond is called psi (y). • The conformation of the mainchain atoms is determined by the values of these two angles for each amino acid. Torsion Angles Phi and Psi Ramachandran • Ramachandran plots indicate allowed combinations of the conformational Plots angles phi and psi. • Since phi (f) and psi (y) refer to rotations of two rigid peptide units around the same Ca atom, most combinations produce steric collisions either between atoms in different peptide groups or between a peptide unit and the side chain attached to Ca. These combinations are therefore not allowed. • Colored areas show sterically allowed regions. The areas labeled a, , and L correspond approximately to conformational angles found for the usual right-handed a helices, strands, and left-handed a helices,respectively. Calculated Ramachandran Plots for Amino Acids Gly with only one H atom as a sidechain, can adopt a much wider range of conformations than the other residues. • (Left) Observed values for all residue types except glycine. Each point represents f and y values for an amino acid residue in a well-refined x-ray structure to high resolution. • (Right) Observed values for glycine. Notice that the values include combinations of f and y that are not allowed for other amino acids. (From J. Richardson, Adv. Prot. Chem. 34: 174175,1981.) Certain Side-chain Conformations are Energetically Favorable 3 conformations of Val • The staggered conformations are the most energetically favored conformations of two tetrahedrally coordinated carbon atoms. Side Chain Conformation • The side chain atoms of amino acids are named using the Greek alphabet according to this scheme. Side Chain Torsion Angles • The side chain torsion angles are named chi1, chi2, chi3, etc., as shown below for lysine. Chi1(χ1) Angles • The chi1 angle is subject to certain restrictions, which arise from steric hindrance between the gamma side chain atom(s) and the main chain. • The different conformations of the side chain as a function of chi1 are referred to as gauche(+), trans and gauche(-). These are indicated in the diagrams here, in which the amino acid is viewed along the C-Ca bond. The most abundant conformation is gauche(+), in which the gamma side chain atom is opposite to the residue's main chain carbonyl group when viewed along the C-Ca bond. Gauche The second most abundant conformation is trans, in which the side chain gamma atom is opposite the main chain nitrogen. The least abundant conformation is gauche(-), which occurs when the side chain is opposite the hydrogen substituent on the Ca atom. This conformation is unstable because the gamma atom is in close contact with the main chain CO and NH groups. The gauche(-) conformation is occasionally adopted by Ser or Thr residues in a helices. Chi2 (2) • In general, side chains tend to adopt the same three torsion angles (+/- 60 and 180 degrees) about chi2 since these correspond to staggered conformations. • However, for residues with an sp2 hybridized gamma atom such as Phe, Tyr, etc., chi2 rarely equals 180 degrees because this would involve an eclipsed conformation. For these side chains the chi2 angle is usually close to +/- 90 degrees as this minimizes close contacts. • For residues such as Asp and Asn the chi2 angles are strongly influenced by the hydrogen bonding capacity of the side chain and its environment. Consequently, these residues adopt a wide range of chi2 angles.