Document
... • The idea of a topology in high dimensional gene expression spaces is not exactly obvious – How do we know what topologies are appropriate? – In practice people often choose nearly square grids for no particularly good reason ...
... • The idea of a topology in high dimensional gene expression spaces is not exactly obvious – How do we know what topologies are appropriate? – In practice people often choose nearly square grids for no particularly good reason ...
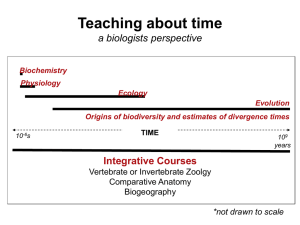
Teaching deep time through macroevolution and
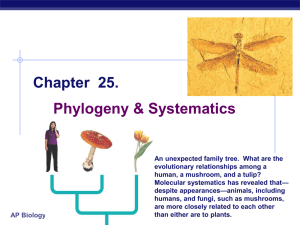
... “tree” based on the similarity of characters--this is done by hand. [cladograms are visual representations of calculated relationships] 2. Students create character matrix and extract DNA/ sequence the 28s rRNA gene. [scaled up repetition, base pair differences are empirically determined] 3. Student ...
... “tree” based on the similarity of characters--this is done by hand. [cladograms are visual representations of calculated relationships] 2. Students create character matrix and extract DNA/ sequence the 28s rRNA gene. [scaled up repetition, base pair differences are empirically determined] 3. Student ...
Package `OrderedList` - USTC Open Source Software Mirror
... The data contains a list with three elements: breast, prostate and map. The first two are expression sets of class ExpressionSet taken from the breast cancer study of Huang et al. (2003) and the prostate cancer study of Singh et al. (2002). Both data sets were preprocessed as described in Yang et al ...
... The data contains a list with three elements: breast, prostate and map. The first two are expression sets of class ExpressionSet taken from the breast cancer study of Huang et al. (2003) and the prostate cancer study of Singh et al. (2002). Both data sets were preprocessed as described in Yang et al ...
“Cheaper, Better, Faster” Bacteria TMDLs
... 1. What is Bacteria Source Tracking (BST)? The term “Bacteria Source Tracking” (BST) in connection with water quality management describes the use of various technologies to determine the source of bacteria in a given body of water. Nationally, several procedures for BST in two major categories (bio ...
... 1. What is Bacteria Source Tracking (BST)? The term “Bacteria Source Tracking” (BST) in connection with water quality management describes the use of various technologies to determine the source of bacteria in a given body of water. Nationally, several procedures for BST in two major categories (bio ...
Are 100 enough? Inferring acanthomorph teleost phylogeny using
... taxonomic range relative to those markers. This facilitates the capture of homologous loci that are useful for both old and more recent divergences, a property shared with UCEs [22, 27]. One advantage to AHE, which we utilize here, is the ease of generating reliable alignments due to the paucity of ...
... taxonomic range relative to those markers. This facilitates the capture of homologous loci that are useful for both old and more recent divergences, a property shared with UCEs [22, 27]. One advantage to AHE, which we utilize here, is the ease of generating reliable alignments due to the paucity of ...
Ramamoorthy, Krithika : Critical Review of Methods available for Microarray Data Analysis
... has a log ratio of -1, and a gene expressed at a constant level has a log ratio of 0. One can use log ratios of other bases as well as long as one is consistent (5). Normalization and noise Before data from multiple microarray experiments can be pooled into a single analysis, the data must first be ...
... has a log ratio of -1, and a gene expressed at a constant level has a log ratio of 0. One can use log ratios of other bases as well as long as one is consistent (5). Normalization and noise Before data from multiple microarray experiments can be pooled into a single analysis, the data must first be ...
Notes
... the longest strings in Genome 1 that have one identical match in Genome 2 – Naïve method: O(N2) – Using suffix tree: O(N) ...
... the longest strings in Genome 1 that have one identical match in Genome 2 – Naïve method: O(N2) – Using suffix tree: O(N) ...
An Exhaustive Epistatic SNP Association Analysis on Expanded
... the multiple-testing correction. In the Wellcome Trust Case Control Consortium (WTCCC) data5, for example, there are over 60 billion pairs of SNPs that can be tested for association, requiring a P value of less than 8 3 10213 for significance under Bonferroni correction. One way to address the multi ...
... the multiple-testing correction. In the Wellcome Trust Case Control Consortium (WTCCC) data5, for example, there are over 60 billion pairs of SNPs that can be tested for association, requiring a P value of less than 8 3 10213 for significance under Bonferroni correction. One way to address the multi ...
Analysis of Gene expression data using MATLAB Software
... cumulative density, respectively. Positive signal components are estimated after adjustment of the background components. This background correction ...
... cumulative density, respectively. Positive signal components are estimated after adjustment of the background components. This background correction ...
Origin and evolution of the slime molds (Mycetozoa)
... The Mycetozoa include the cellular (dictyostelid), acellular (myxogastrid), and protostelid slime molds. However, available molecular data are in disagreement on both the monophyly and phylogenetic position of the group. Ribosomal RNA trees show the myxogastrid and dictyostelid slime molds as unrela ...
... The Mycetozoa include the cellular (dictyostelid), acellular (myxogastrid), and protostelid slime molds. However, available molecular data are in disagreement on both the monophyly and phylogenetic position of the group. Ribosomal RNA trees show the myxogastrid and dictyostelid slime molds as unrela ...
No Slide Title - Brigham Young University
... SOMs are unsupervised neural net algorithms that identify coregulated genes Page 211 ...
... SOMs are unsupervised neural net algorithms that identify coregulated genes Page 211 ...
Extraction of correlated gene clusters from multiple genomic data by
... Recent developments in high-throughput technologies have filled biological databases with many kinds of genomic data, such as pathway knowledge (Kanehisa et al., 2002), microarray gene expression data (Eisen et al., 1998), protein-protein interaction data (Ito et al., 2001), phylogenetic profiles (P ...
... Recent developments in high-throughput technologies have filled biological databases with many kinds of genomic data, such as pathway knowledge (Kanehisa et al., 2002), microarray gene expression data (Eisen et al., 1998), protein-protein interaction data (Ito et al., 2001), phylogenetic profiles (P ...
Inked
... 2. The function is: 3. There are ____________ and _______________regions 4. The gene tolerates: B. The gene encoding 16S ribosomal RNA (18S in eukaryotes) 1. The 16S rRNA is part of the ribosome a) Occurs in _______________________ b) c) Gene size: d) composed of many domains that can change indepen ...
... 2. The function is: 3. There are ____________ and _______________regions 4. The gene tolerates: B. The gene encoding 16S ribosomal RNA (18S in eukaryotes) 1. The 16S rRNA is part of the ribosome a) Occurs in _______________________ b) c) Gene size: d) composed of many domains that can change indepen ...
Innovative Database Models and Advanced Tools in Bioinformatics
... parameters. Searching data using optimal alignment algorithms, rather than using heuristic methods. Giving researchers/clinicians the ability to formulate sequence identification concepts and test their ideas against a validated database Incorporating information from GenBank if needed ...
... parameters. Searching data using optimal alignment algorithms, rather than using heuristic methods. Giving researchers/clinicians the ability to formulate sequence identification concepts and test their ideas against a validated database Incorporating information from GenBank if needed ...
Package `DART`
... (3) PruneNet: This function obtains the pruned, i.e consistent, network, in which any edge represents a significant correlation in gene expression whose directionality agrees with that predicted by the prior information. This is the denoising step of the algorithm. The function returns the whole pru ...
... (3) PruneNet: This function obtains the pruned, i.e consistent, network, in which any edge represents a significant correlation in gene expression whose directionality agrees with that predicted by the prior information. This is the denoising step of the algorithm. The function returns the whole pru ...
The Combination of Genetic Programming and Genetic Algorithm for
... algorithm that represent the networks as two dimensional array of binary digit . After that in 1990 Kitan seen the process of direct encoding of array is be more complex when the design is big , he suggest a grammatical encoding for network by using some of grammatical rules see [Mit. 96] . Genetic ...
... algorithm that represent the networks as two dimensional array of binary digit . After that in 1990 Kitan seen the process of direct encoding of array is be more complex when the design is big , he suggest a grammatical encoding for network by using some of grammatical rules see [Mit. 96] . Genetic ...
cluster analysis I
... 1. EM algorithm to maximize the mixture likelihood. 2. Model selection: Bayesian Information Criterion (BIC) for determining k and the complexity of the covariance matrix. ...
... 1. EM algorithm to maximize the mixture likelihood. 2. Model selection: Bayesian Information Criterion (BIC) for determining k and the complexity of the covariance matrix. ...
Association Test with the Principal Component Analysis in Case
... components and is asymptotically distributed as a c 2 distribution with one degree of freedom. Simulation studies showed that, when the number of markers is not very large, the proposed statistic has higher power than the APRICOT test which is based on the same method of the principal-component anal ...
... components and is asymptotically distributed as a c 2 distribution with one degree of freedom. Simulation studies showed that, when the number of markers is not very large, the proposed statistic has higher power than the APRICOT test which is based on the same method of the principal-component anal ...
The Combination of Genetic Programming and Genetic Algorithm for
... network by some rules as follow : 1- if the problem that represented by the neural network have one output neurons , then the first node in tree it’s output node .Else (much than 2 neurons) the second level of tree represented to be the output layer and the first node is just be root node no more . ...
... network by some rules as follow : 1- if the problem that represented by the neural network have one output neurons , then the first node in tree it’s output node .Else (much than 2 neurons) the second level of tree represented to be the output layer and the first node is just be root node no more . ...
Segway: simultaneous segmentation of multiple functional genomics
... The method partitions the genome into variable-length segments using a dynamic Bayesian network where the dynamic (or “time”) axis represents genomic position. Segments are assigned one of a finite number of labels such that the vectors of observations are similar in segments with the same label. A ...
... The method partitions the genome into variable-length segments using a dynamic Bayesian network where the dynamic (or “time”) axis represents genomic position. Segments are assigned one of a finite number of labels such that the vectors of observations are similar in segments with the same label. A ...
Clustered alignments of gene-expression time series data
... – a dynamic programming algorithm designed to find an optimal alignment between two series with multiple channels of information(such as genes). – Briefly, it aligns and scores two give time series based on their similarity – Two series as q (for query series) and d (for database series) – The serie ...
... – a dynamic programming algorithm designed to find an optimal alignment between two series with multiple channels of information(such as genes). – Briefly, it aligns and scores two give time series based on their similarity – Two series as q (for query series) and d (for database series) – The serie ...
Clustering_PartII_2012
... Orientation of the nodes is irrelevant …. although some clustering programs try to organize nodes in some way. ...
... Orientation of the nodes is irrelevant …. although some clustering programs try to organize nodes in some way. ...